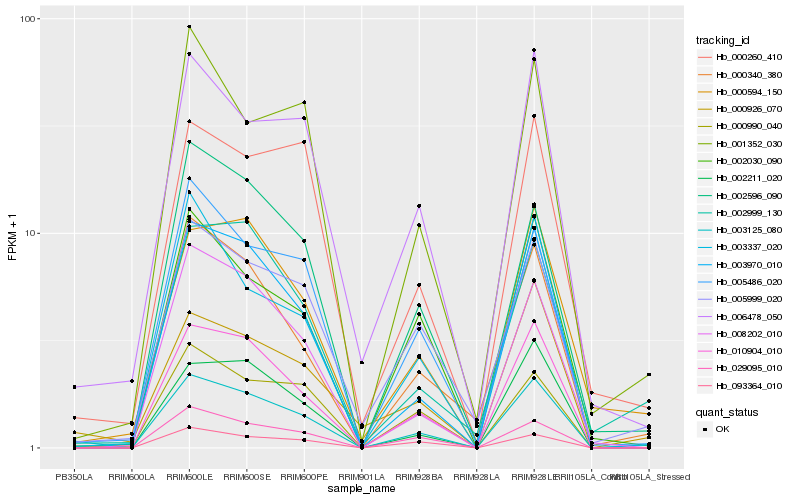
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003125_080 |
0.0 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 2 |
Hb_010904_010 |
0.1314691782 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003970_010 |
0.1406839798 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 4 |
Hb_000926_070 |
0.1426041272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006478_050 |
0.1491533247 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005486_020 |
0.1494776032 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 7 |
Hb_029095_010 |
0.1521093727 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 8 |
Hb_000340_380 |
0.1539485193 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 9 |
Hb_002999_130 |
0.1545754098 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 10 |
Hb_005999_020 |
0.1548130892 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 11 |
Hb_000594_150 |
0.1549437146 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 12 |
Hb_000990_040 |
0.1550703671 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 13 |
Hb_002211_020 |
0.1572302813 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 14 |
Hb_002596_090 |
0.157354045 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_008202_010 |
0.1589668823 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_002030_090 |
0.1589992234 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 17 |
Hb_001352_030 |
0.1590854057 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 18 |
Hb_003337_020 |
0.1600273914 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 19 |
Hb_000260_410 |
0.1626644275 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 20 |
Hb_093364_010 |
0.1627192954 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |