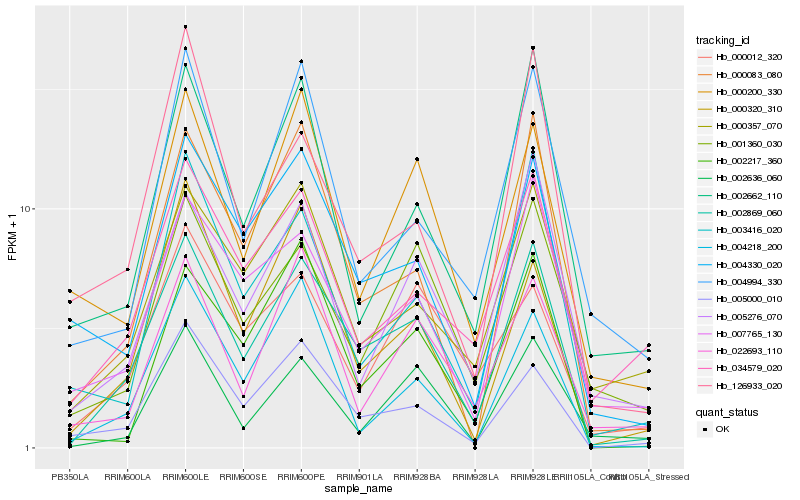
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002869_060 |
0.0 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 2 |
Hb_005000_010 |
0.14184203 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000083_080 |
0.1444581858 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 4 |
Hb_007765_130 |
0.1531684162 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004330_020 |
0.156722459 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 6 |
Hb_005276_070 |
0.1618905451 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 7 |
Hb_000320_310 |
0.1624423162 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |
| 8 |
Hb_001360_030 |
0.1634695881 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 9 |
Hb_000357_070 |
0.1635709812 |
- |
- |
- |
| 10 |
Hb_002217_360 |
0.1637066986 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_034579_020 |
0.1657974076 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002662_110 |
0.1662921461 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 13 |
Hb_003416_020 |
0.1665193189 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 14 |
Hb_002636_060 |
0.1704128767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004218_200 |
0.1706035078 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 16 |
Hb_000200_330 |
0.1727759391 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 17 |
Hb_004994_330 |
0.1735402112 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_022693_110 |
0.1774725278 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 19 |
Hb_000012_320 |
0.1779672976 |
- |
- |
PREDICTED: uncharacterized protein LOC105638164 [Jatropha curcas] |
| 20 |
Hb_126933_020 |
0.179017637 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |