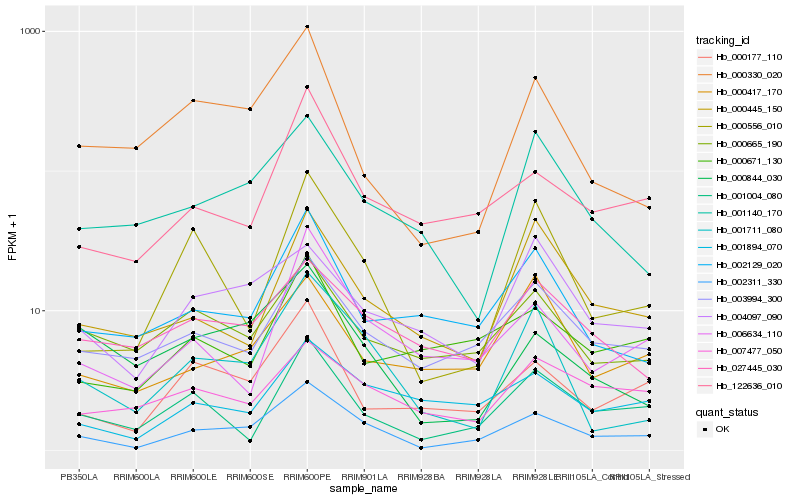
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_330 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_003994_300 |
0.174549489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_122636_010 |
0.1749847483 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 4 |
Hb_000177_110 |
0.1778450192 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 5 |
Hb_001711_080 |
0.1839959662 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 6 |
Hb_001894_070 |
0.1900856407 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006634_110 |
0.1926496793 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 8 |
Hb_000665_190 |
0.1988082739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001004_080 |
0.2001807421 |
- |
- |
hypothetical protein POPTR_0014s14130g [Populus trichocarpa] |
| 10 |
Hb_002129_020 |
0.2008314018 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 11 |
Hb_027445_030 |
0.2040592714 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_000844_030 |
0.2059863066 |
- |
- |
ent-kaurene oxidase, putative [Ricinus communis] |
| 13 |
Hb_000330_020 |
0.2064301372 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 14 |
Hb_000556_010 |
0.2064777918 |
- |
- |
glycerate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_004097_090 |
0.2071400758 |
- |
- |
PREDICTED: uncharacterized protein LOC105634517 [Jatropha curcas] |
| 16 |
Hb_001140_170 |
0.209568813 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 17 |
Hb_007477_050 |
0.2096232859 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000445_150 |
0.2108366175 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 19 |
Hb_000671_130 |
0.2114794674 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000417_170 |
0.2116975562 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |