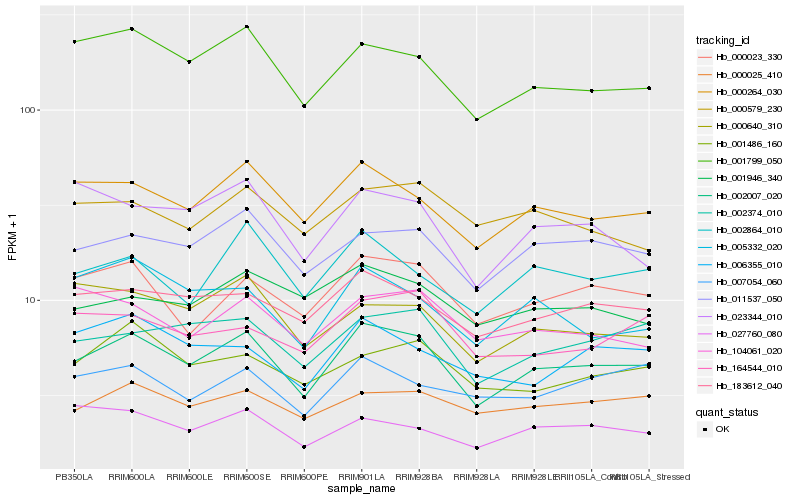
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002007_020 |
0.0 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like SIM [Jatropha curcas] |
| 2 |
Hb_000264_030 |
0.0746953798 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 3 |
Hb_011537_050 |
0.0795241883 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001799_050 |
0.0796750623 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 5 |
Hb_000023_330 |
0.0822954283 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 6 |
Hb_183612_040 |
0.0833397324 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 7 |
Hb_002374_010 |
0.0844437718 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002864_010 |
0.0908172147 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 9 |
Hb_000025_410 |
0.095733282 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g01110-like [Populus euphratica] |
| 10 |
Hb_001486_160 |
0.0991476567 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 11 |
Hb_001946_340 |
0.1006435212 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_164544_010 |
0.1016138503 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.5 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000640_310 |
0.1017424269 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 14 |
Hb_006355_010 |
0.1021112726 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 15 |
Hb_027760_080 |
0.1021347718 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_005332_020 |
0.1026183895 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 17 |
Hb_000579_230 |
0.1031295166 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 18 |
Hb_023344_010 |
0.1031429721 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007054_060 |
0.1036144386 |
- |
- |
- |
| 20 |
Hb_104061_020 |
0.1037720435 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |