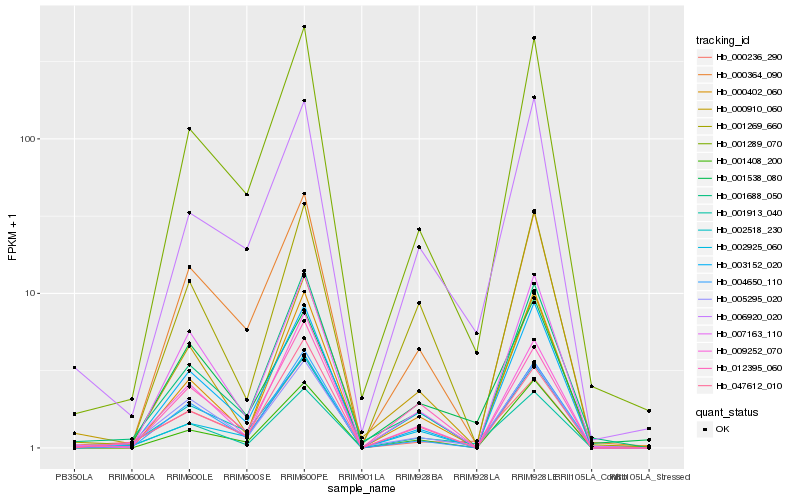
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001913_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005295_020 |
0.1124816876 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 3 |
Hb_009252_070 |
0.129911485 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 4 |
Hb_001269_660 |
0.1300955041 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 5 |
Hb_001538_080 |
0.1320349617 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_006920_020 |
0.1365476391 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 7 |
Hb_001289_070 |
0.1379922709 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 8 |
Hb_002925_060 |
0.1382144006 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 9 |
Hb_003152_020 |
0.1390900542 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 10 |
Hb_007163_110 |
0.1399954027 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 11 |
Hb_047612_010 |
0.1415406507 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 12 |
Hb_000910_060 |
0.1431581792 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_000236_290 |
0.1440003499 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 14 |
Hb_002518_230 |
0.1443823484 |
- |
- |
- |
| 15 |
Hb_004650_110 |
0.1455971796 |
- |
- |
hypothetical protein POPTR_0002s08760g [Populus trichocarpa] |
| 16 |
Hb_012395_060 |
0.147179924 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 17 |
Hb_000402_060 |
0.147228128 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001408_200 |
0.1496716053 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 19 |
Hb_001688_050 |
0.1501251972 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 20 |
Hb_000364_090 |
0.1503761436 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X2 [Jatropha curcas] |