| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
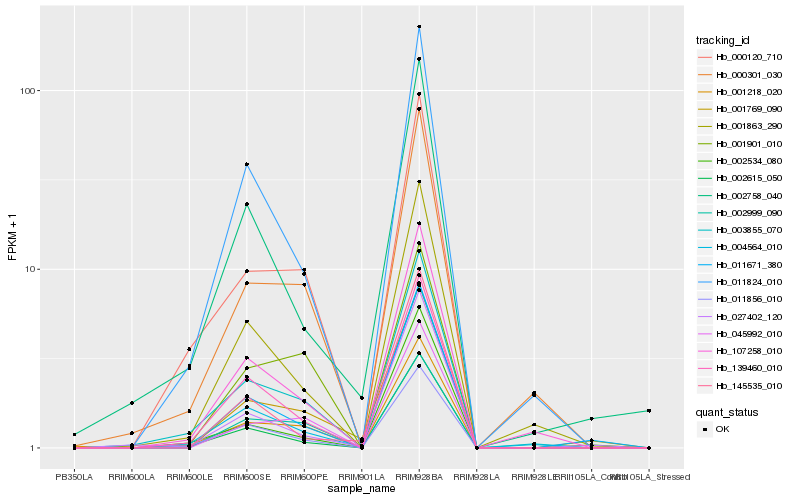
Hb_001769_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630842 [Jatropha curcas] |
| 2 |
Hb_011671_380 |
0.1037899634 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 3 |
Hb_045992_010 |
0.1240683916 |
- |
- |
- |
| 4 |
Hb_027402_120 |
0.1386833458 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_001218_020 |
0.1413074969 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s17080g [Populus trichocarpa] |
| 6 |
Hb_002999_090 |
0.1463829548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_139460_010 |
0.1489636805 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 8 |
Hb_000301_030 |
0.1497356696 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 9 |
Hb_145535_010 |
0.1504029567 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 10 |
Hb_003855_070 |
0.1520917347 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 11 |
Hb_011856_010 |
0.1524029283 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_001901_010 |
0.1524255914 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 13 |
Hb_107258_010 |
0.1541343639 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 14 |
Hb_001863_290 |
0.1544389238 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 15 |
Hb_002615_050 |
0.1547472358 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 16 |
Hb_004564_010 |
0.1562989925 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 17 |
Hb_002758_040 |
0.1569581124 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000120_710 |
0.1571494242 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_002534_080 |
0.1587638903 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_011824_010 |
0.1590351546 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |