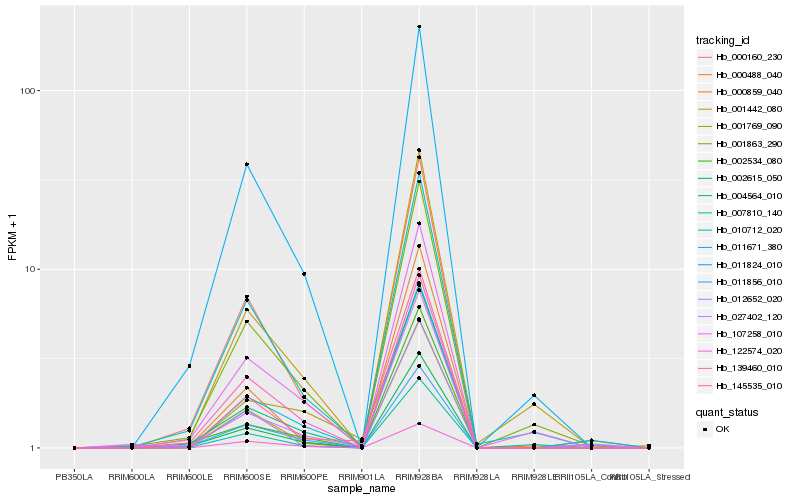
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_380 |
0.0 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 2 |
Hb_027402_120 |
0.0996982075 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_001769_090 |
0.1037899634 |
- |
- |
PREDICTED: uncharacterized protein LOC105630842 [Jatropha curcas] |
| 4 |
Hb_002615_050 |
0.1038530316 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 5 |
Hb_007810_140 |
0.1066569342 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 6 |
Hb_139460_010 |
0.1082138764 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 7 |
Hb_001863_290 |
0.1085300264 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 8 |
Hb_002534_080 |
0.1107373706 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000160_230 |
0.1109826449 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 10 |
Hb_011824_010 |
0.1124825672 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_000859_040 |
0.1127755809 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 12 |
Hb_004564_010 |
0.1182627763 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 13 |
Hb_011856_010 |
0.1196712562 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_107258_010 |
0.1200101586 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 15 |
Hb_122574_020 |
0.1242704167 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 16 |
Hb_000488_040 |
0.1251126004 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_010712_020 |
0.1255717472 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 18 |
Hb_001442_080 |
0.1264120753 |
transcription factor |
TF Family: MYB-related |
transcription factor MYB251 [Fagus crenata] |
| 19 |
Hb_145535_010 |
0.1264291108 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 20 |
Hb_012652_020 |
0.1266749164 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |