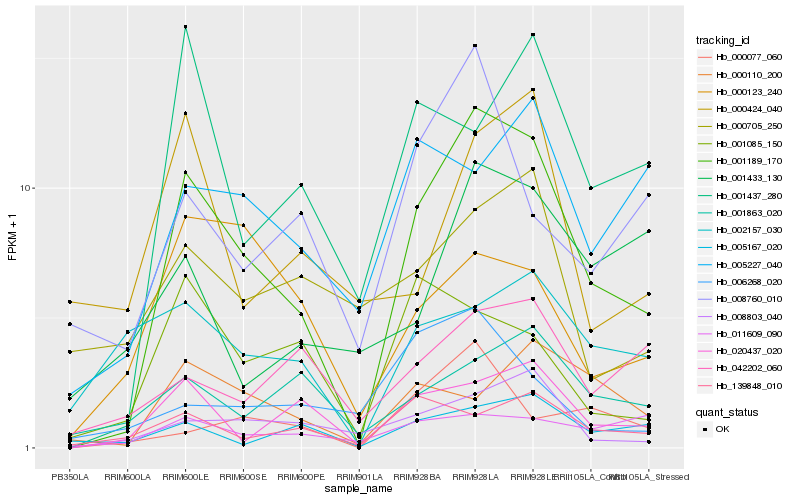
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001189_170 |
0.0 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_1340820 [Ricinus communis] |
| 2 |
Hb_020437_020 |
0.2355664417 |
- |
- |
hypothetical protein POPTR_0001s01410g, partial [Populus trichocarpa] |
| 3 |
Hb_008803_040 |
0.2524010145 |
- |
- |
PREDICTED: uncharacterized protein LOC105157106 [Sesamum indicum] |
| 4 |
Hb_001437_280 |
0.2624692785 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 5 |
Hb_139848_010 |
0.2696726616 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS4, chloroplastic [Populus euphratica] |
| 6 |
Hb_000110_200 |
0.2731756578 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_005167_020 |
0.2736833539 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 8 |
Hb_001863_020 |
0.2772007569 |
- |
- |
PREDICTED: protein FAM91A1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005227_040 |
0.2809751044 |
transcription factor |
TF Family: HB |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_008760_010 |
0.2816823963 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 11 |
Hb_001085_150 |
0.2820985568 |
- |
- |
Purple acid phosphatase 15 [Theobroma cacao] |
| 12 |
Hb_011609_090 |
0.2856048956 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000077_060 |
0.2866331654 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 14 |
Hb_042202_060 |
0.2927411396 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Jatropha curcas] |
| 15 |
Hb_002157_030 |
0.2930797557 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000123_240 |
0.2936903285 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 17 |
Hb_000424_040 |
0.2937359034 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 18 |
Hb_000705_250 |
0.2963374023 |
- |
- |
hypothetical protein CICLE_v10017607mg [Citrus clementina] |
| 19 |
Hb_001433_130 |
0.2976674972 |
- |
- |
PREDICTED: uncharacterized protein LOC105647468 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006268_020 |
0.3002979412 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |