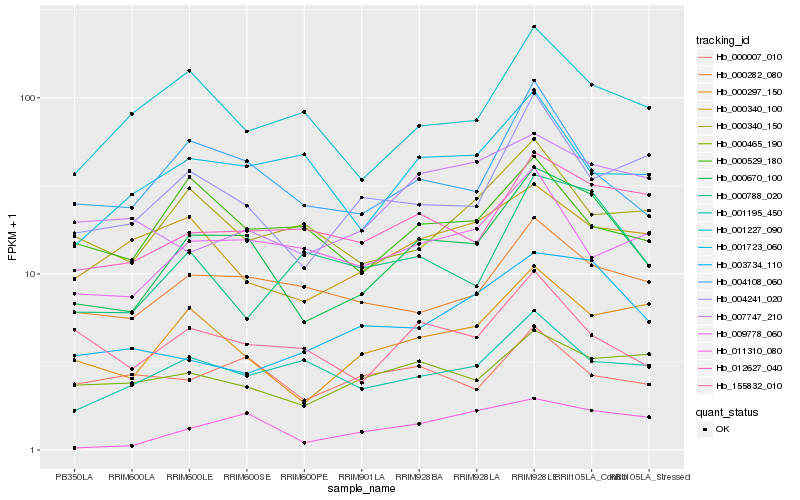
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000670_100 |
0.0 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 3, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_004108_060 |
0.1478523728 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 3 |
Hb_000297_150 |
0.1505247858 |
- |
- |
PREDICTED: uncharacterized protein LOC103500908 [Cucumis melo] |
| 4 |
Hb_012627_040 |
0.1553912514 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 5 |
Hb_000282_080 |
0.1602708999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004241_020 |
0.1677263893 |
- |
- |
PREDICTED: DAG protein, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_011310_080 |
0.1717581447 |
- |
- |
hypothetical protein POPTR_0006s26840g [Populus trichocarpa] |
| 8 |
Hb_001227_090 |
0.1729982301 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 9 |
Hb_000465_190 |
0.1748998644 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_155832_010 |
0.1750703103 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 11 |
Hb_000788_020 |
0.1764305228 |
- |
- |
PREDICTED: uncharacterized protein LOC105629709 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009778_060 |
0.1764585587 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 13 |
Hb_000340_100 |
0.1765693574 |
- |
- |
PREDICTED: protein PALE CRESS, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001195_450 |
0.1771675207 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 15 |
Hb_000340_150 |
0.182886316 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 16 |
Hb_000529_180 |
0.1831700654 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001723_060 |
0.1841571376 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 18 |
Hb_007747_210 |
0.1850525782 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_003734_110 |
0.1851758286 |
- |
- |
PREDICTED: cytochrome c-type biogenesis ccda-like chloroplastic protein [Jatropha curcas] |
| 20 |
Hb_000007_010 |
0.1894443325 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |