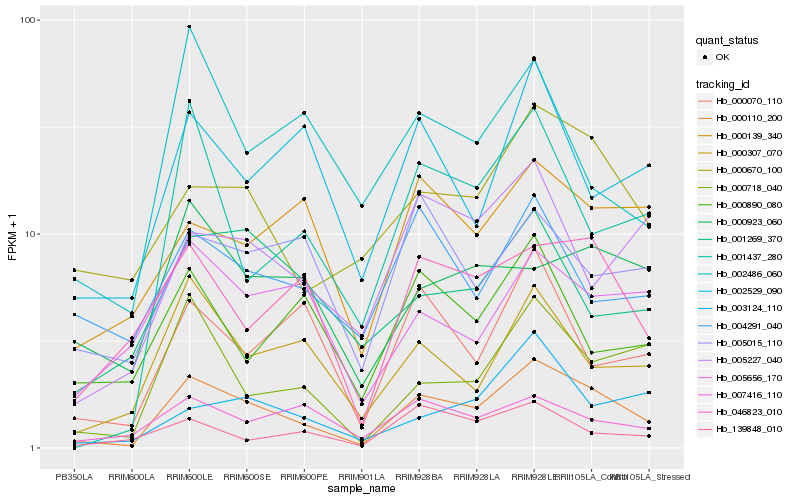
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_200 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000718_040 |
0.1927818348 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 3 |
Hb_000670_100 |
0.2000537055 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 3, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000307_070 |
0.2014965472 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 5 |
Hb_046823_010 |
0.2049556427 |
- |
- |
hypothetical protein CICLE_v100021301mg, partial [Citrus clementina] |
| 6 |
Hb_001437_280 |
0.2095008925 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 7 |
Hb_000070_110 |
0.2169249571 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007416_110 |
0.2173074922 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002529_090 |
0.2202396168 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_005227_040 |
0.2212811636 |
transcription factor |
TF Family: HB |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_001269_370 |
0.2241637969 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 12 |
Hb_000139_340 |
0.2253169817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005656_170 |
0.2261284219 |
- |
- |
PREDICTED: uncharacterized protein LOC105637447 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005015_110 |
0.2270522454 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 15 |
Hb_004291_040 |
0.2278414104 |
- |
- |
hexokinase [Manihot esculenta] |
| 16 |
Hb_003124_110 |
0.2280242266 |
- |
- |
hypothetical protein VITISV_026680 [Vitis vinifera] |
| 17 |
Hb_139848_010 |
0.2310904842 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS4, chloroplastic [Populus euphratica] |
| 18 |
Hb_000890_080 |
0.2314149882 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 19 |
Hb_000923_060 |
0.2325283507 |
- |
- |
PREDICTED: uncharacterized protein LOC105628271 [Jatropha curcas] |
| 20 |
Hb_002486_060 |
0.2331776393 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |