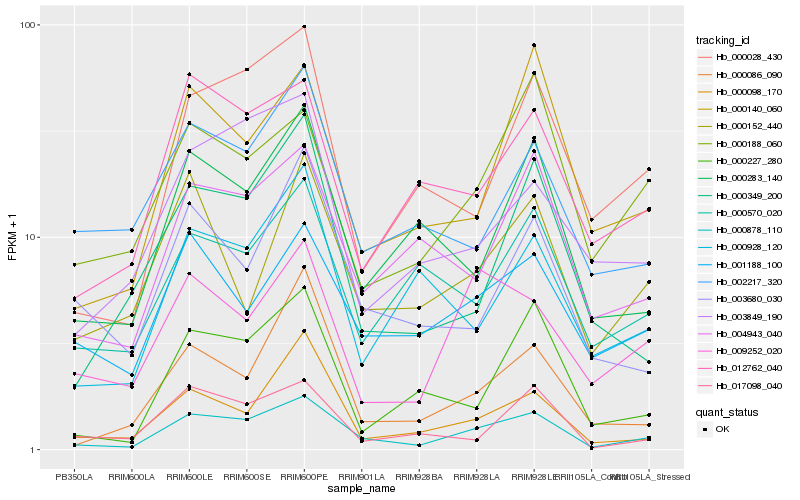
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000878_110 |
0.0 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 2 |
Hb_004943_040 |
0.1738261516 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000028_430 |
0.1767867915 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001188_100 |
0.1777509101 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 5 |
Hb_000283_140 |
0.1800780305 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000140_060 |
0.1801015967 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 7 |
Hb_012762_040 |
0.1803728343 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 8 |
Hb_000098_170 |
0.1821155171 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 9 |
Hb_000227_280 |
0.1825545371 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 10 |
Hb_009252_020 |
0.1837117797 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 11 |
Hb_017098_040 |
0.1899937701 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 12 |
Hb_000188_060 |
0.1901825013 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 13 |
Hb_000086_090 |
0.1904983208 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 14 |
Hb_000570_020 |
0.1913101441 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 15 |
Hb_000349_200 |
0.1914247156 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g12500 [Jatropha curcas] |
| 16 |
Hb_003680_030 |
0.1938875756 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000152_440 |
0.19568255 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 18 |
Hb_002217_320 |
0.19587422 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000928_120 |
0.1959058511 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 20 |
Hb_003849_190 |
0.1959162135 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |