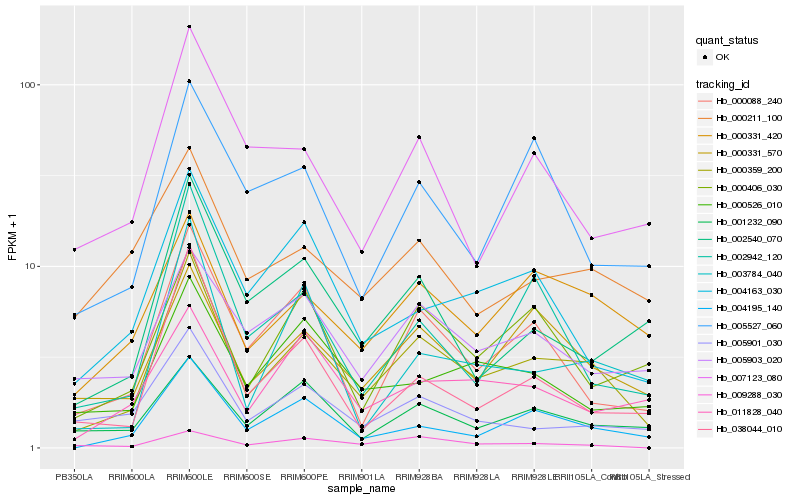
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_200 |
0.0 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 2 |
Hb_004195_140 |
0.1564746309 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 3 |
Hb_000331_570 |
0.1602410751 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 4 |
Hb_000088_240 |
0.1631867215 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000331_420 |
0.1642992424 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001232_090 |
0.1745726006 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 7 |
Hb_038044_010 |
0.1814754737 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 8 |
Hb_000406_030 |
0.1817425729 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 9 |
Hb_002942_120 |
0.1817748056 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 10 |
Hb_004163_030 |
0.1832175635 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 11 |
Hb_005901_030 |
0.1838051771 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005903_020 |
0.184650393 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 13 |
Hb_009288_030 |
0.1852876661 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 14 |
Hb_002540_070 |
0.1857160348 |
- |
- |
PREDICTED: protein cornichon homolog 1 [Jatropha curcas] |
| 15 |
Hb_007123_080 |
0.1874797146 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 16 |
Hb_000526_010 |
0.1877681494 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 17 |
Hb_003784_040 |
0.1908845135 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 18 |
Hb_005527_060 |
0.1909792438 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 19 |
Hb_000211_100 |
0.191649481 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 20 |
Hb_011828_040 |
0.1920190218 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |