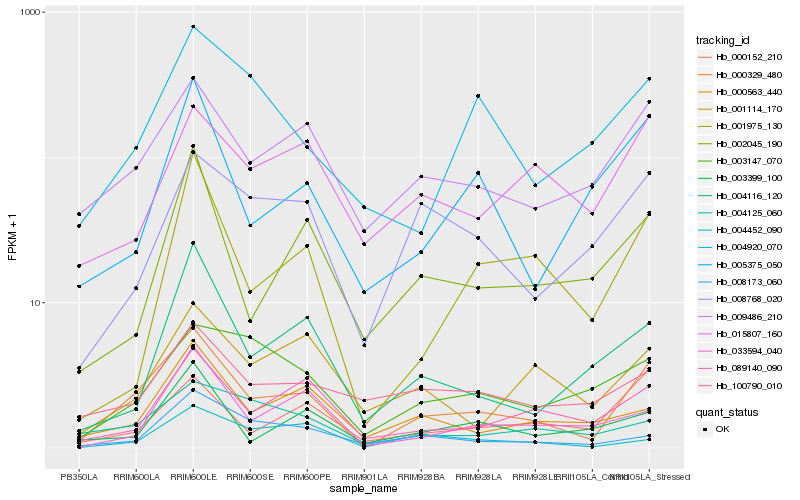
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_480 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 4 [Jatropha curcas] |
| 2 |
Hb_009486_210 |
0.2010837756 |
- |
- |
- |
| 3 |
Hb_089140_090 |
0.22549478 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000563_440 |
0.2376837365 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000152_210 |
0.2389477814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008768_020 |
0.2437471058 |
- |
- |
PREDICTED: uncharacterized protein LOC105639378 [Jatropha curcas] |
| 7 |
Hb_005375_050 |
0.2440713644 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 8 |
Hb_002045_190 |
0.2441839764 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 9 |
Hb_033594_040 |
0.2442103103 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_001975_130 |
0.2448485352 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 6 isoform X1 [Populus euphratica] |
| 11 |
Hb_004125_060 |
0.2461434788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004116_120 |
0.2464925023 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 13 |
Hb_004920_070 |
0.2476601421 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 14 |
Hb_008173_060 |
0.249450618 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 15 |
Hb_001114_170 |
0.2504300846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003147_070 |
0.2511824728 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 17 |
Hb_100790_010 |
0.2528617868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_015807_160 |
0.253684327 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 19 |
Hb_004452_090 |
0.2544557753 |
- |
- |
hypothetical protein JCGZ_12231 [Jatropha curcas] |
| 20 |
Hb_003399_100 |
0.2547878941 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |