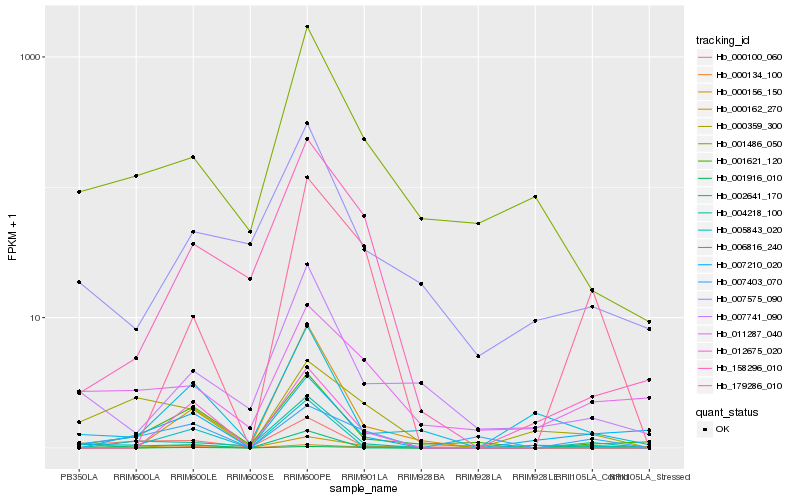
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000162_270 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104442671 [Eucalyptus grandis] |
| 2 |
Hb_007403_070 |
0.2523420406 |
- |
- |
hypothetical protein JCGZ_23672 [Jatropha curcas] |
| 3 |
Hb_179286_010 |
0.2533162788 |
- |
- |
STRUWWELPETER family protein [Populus trichocarpa] |
| 4 |
Hb_004218_100 |
0.2675161953 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 5C [Jatropha curcas] |
| 5 |
Hb_001621_120 |
0.2830727842 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 6 |
Hb_005843_020 |
0.2844453231 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 7 |
Hb_000359_300 |
0.2866890944 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Zea mays] |
| 8 |
Hb_002641_170 |
0.2882044574 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_011287_040 |
0.2952280986 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007210_020 |
0.2957782502 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial-like [Solanum tuberosum] |
| 11 |
Hb_158296_010 |
0.2976606608 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 12 |
Hb_001486_050 |
0.3007477539 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 13 |
Hb_000134_100 |
0.3037749168 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Prunus mume] |
| 14 |
Hb_000100_060 |
0.3085781395 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 15 |
Hb_006816_240 |
0.3095879114 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 16 |
Hb_007741_090 |
0.3101757626 |
- |
- |
Autophagy-related 8f -like protein [Gossypium arboreum] |
| 17 |
Hb_012675_020 |
0.313138757 |
- |
- |
hypothetical protein CISIN_1g018227mg [Citrus sinensis] |
| 18 |
Hb_001916_010 |
0.3134389616 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 19 |
Hb_007575_090 |
0.3156359429 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 20 |
Hb_000156_150 |
0.3166523071 |
- |
- |
hypothetical protein JCGZ_04603 [Jatropha curcas] |