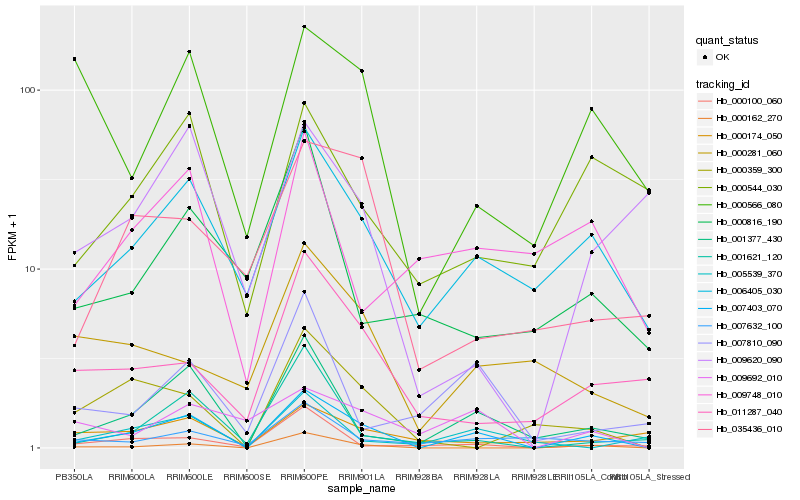
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007403_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_23672 [Jatropha curcas] |
| 2 |
Hb_001377_430 |
0.239204362 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 3 |
Hb_000162_270 |
0.2523420406 |
- |
- |
PREDICTED: uncharacterized protein LOC104442671 [Eucalyptus grandis] |
| 4 |
Hb_006405_030 |
0.2528912137 |
- |
- |
hypothetical protein VITISV_043238 [Vitis vinifera] |
| 5 |
Hb_001621_120 |
0.2940583831 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 6 |
Hb_000100_060 |
0.3027490122 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 7 |
Hb_000359_300 |
0.3114324568 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Zea mays] |
| 8 |
Hb_011287_040 |
0.3177150759 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_009692_010 |
0.3253855639 |
- |
- |
- |
| 10 |
Hb_007810_090 |
0.3257671969 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 11 |
Hb_007632_100 |
0.3264883266 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 12 |
Hb_000566_080 |
0.329048191 |
- |
- |
- |
| 13 |
Hb_035436_010 |
0.3300644781 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 14 |
Hb_009748_010 |
0.3302729604 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 15 |
Hb_005539_370 |
0.3307615239 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_000281_060 |
0.3322154001 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000544_030 |
0.3350058821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000174_050 |
0.3366161877 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 [Populus euphratica] |
| 19 |
Hb_009620_090 |
0.3382343292 |
- |
- |
hypothetical protein L484_000980 [Morus notabilis] |
| 20 |
Hb_000816_190 |
0.3408146218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |