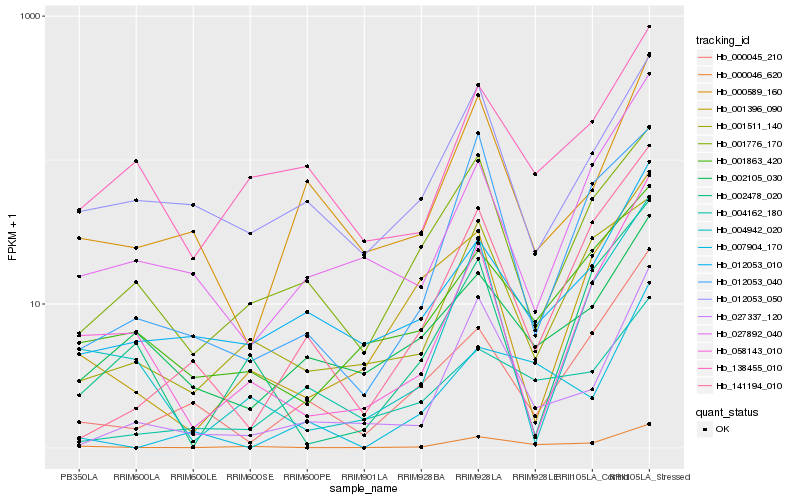
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_620 |
0.0 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 2 |
Hb_001863_420 |
0.1754208013 |
- |
- |
PREDICTED: probable protein phosphatase 2C 10, partial [Vitis vinifera] |
| 3 |
Hb_138455_010 |
0.1842941551 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_012053_010 |
0.186667402 |
- |
- |
srpk, putative [Ricinus communis] |
| 5 |
Hb_027337_120 |
0.1912785255 |
- |
- |
PREDICTED: uncharacterized protein LOC104430249 [Eucalyptus grandis] |
| 6 |
Hb_058143_010 |
0.1979604993 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 7 |
Hb_001511_140 |
0.1998998509 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 8 |
Hb_004942_020 |
0.2000738187 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 9 |
Hb_001776_170 |
0.212021614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_027892_040 |
0.2124539713 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 11 |
Hb_000045_210 |
0.2147238791 |
- |
- |
- |
| 12 |
Hb_141194_010 |
0.21693727 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR 1-like isoform X2 [Populus euphratica] |
| 13 |
Hb_002105_030 |
0.2175983139 |
- |
- |
- |
| 14 |
Hb_012053_050 |
0.2179521783 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 15 |
Hb_007904_170 |
0.2191219015 |
- |
- |
PREDICTED: anther-specific protein BCP1 [Jatropha curcas] |
| 16 |
Hb_001396_090 |
0.2216459562 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 17 |
Hb_000589_160 |
0.2242306421 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 18 |
Hb_004162_180 |
0.2247967089 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_012053_040 |
0.2255167648 |
- |
- |
PREDICTED: putative phospholipid:diacylglycerol acyltransferase 2 [Jatropha curcas] |
| 20 |
Hb_002478_020 |
0.2278732822 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |