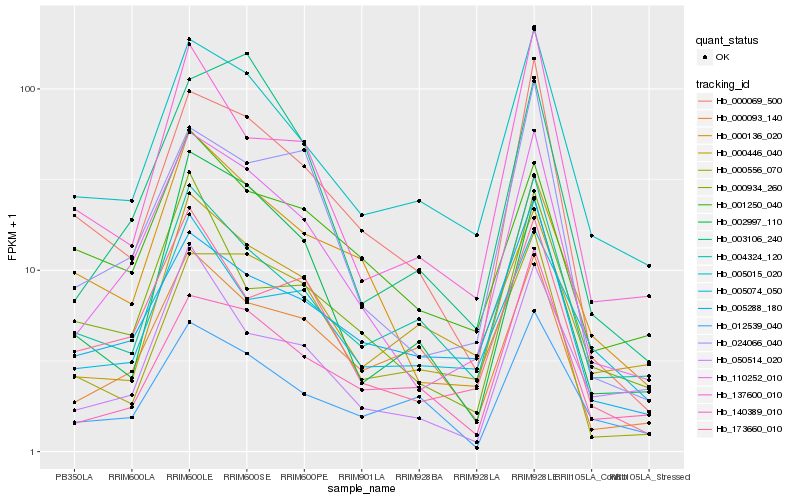
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_110252_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_16705 [Jatropha curcas] |
| 2 |
Hb_005015_020 |
0.1243431488 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_173660_010 |
0.1380082345 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Malus domestica] |
| 4 |
Hb_000069_500 |
0.1409186054 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 4, chloroplastic [Jatropha curcas] |
| 5 |
Hb_024066_040 |
0.142916002 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 6 |
Hb_005288_180 |
0.1572810215 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000136_020 |
0.1580244791 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g80440-like [Jatropha curcas] |
| 8 |
Hb_004324_120 |
0.1608945939 |
- |
- |
ATP-dependent zinc metalloprotease YME1L1 [Gossypium arboreum] |
| 9 |
Hb_137600_010 |
0.161252514 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 10 |
Hb_003106_240 |
0.1624768671 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 11 |
Hb_000093_140 |
0.1632910793 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_050514_020 |
0.1665064762 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 13 |
Hb_140389_010 |
0.1699644864 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 14 |
Hb_000556_070 |
0.1719308594 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At2g45590 [Jatropha curcas] |
| 15 |
Hb_000446_040 |
0.1719397077 |
- |
- |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 16 |
Hb_000934_260 |
0.173699244 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001250_040 |
0.1760275799 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 18 |
Hb_012539_040 |
0.1794475447 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_07765 [Jatropha curcas] |
| 19 |
Hb_002997_110 |
0.1797464704 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005074_050 |
0.1818878873 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |