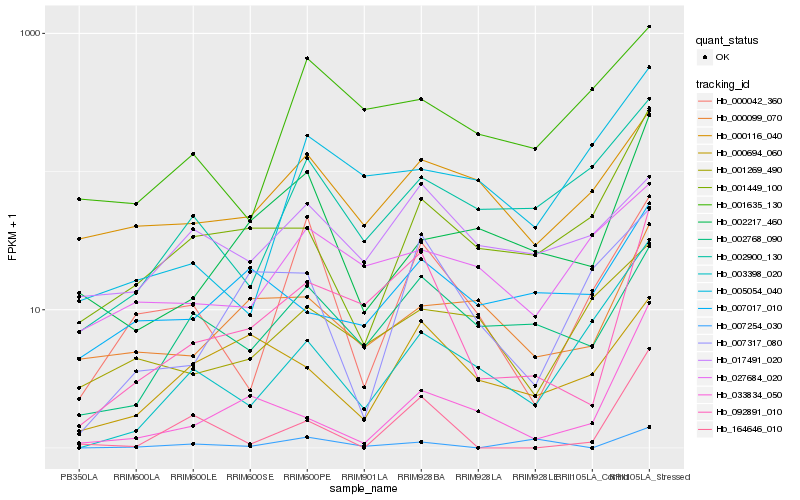
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092891_010 |
0.0 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 2 |
Hb_002768_090 |
0.2425280125 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 3 |
Hb_000099_070 |
0.274159336 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 4 |
Hb_001449_100 |
0.2826391031 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 5 |
Hb_000116_040 |
0.2827879522 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000694_060 |
0.2867500601 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 7 |
Hb_007017_010 |
0.2909234277 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 8 |
Hb_002217_460 |
0.2943074201 |
- |
- |
PREDICTED: transcription factor bHLH147-like [Jatropha curcas] |
| 9 |
Hb_033834_050 |
0.29717973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001635_130 |
0.2987049046 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 11 |
Hb_000042_360 |
0.2987297983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_017491_020 |
0.2993417492 |
- |
- |
hypothetical protein B456_011G056700 [Gossypium raimondii] |
| 13 |
Hb_002900_130 |
0.3003703862 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 14 |
Hb_027684_020 |
0.3044835092 |
- |
- |
ribosomal protein S10 (mitochondrion) [Nicotiana tabacum] |
| 15 |
Hb_007317_080 |
0.3062019995 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 16 |
Hb_005054_040 |
0.3073782238 |
- |
- |
PREDICTED: 60S ribosomal protein L35a-1 [Gossypium raimondii] |
| 17 |
Hb_001269_490 |
0.3076038958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007254_030 |
0.3098493725 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_003398_020 |
0.3101602255 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 20 |
Hb_164646_010 |
0.312429778 |
- |
- |
PREDICTED: acidic endochitinase-like [Pyrus x bretschneideri] |