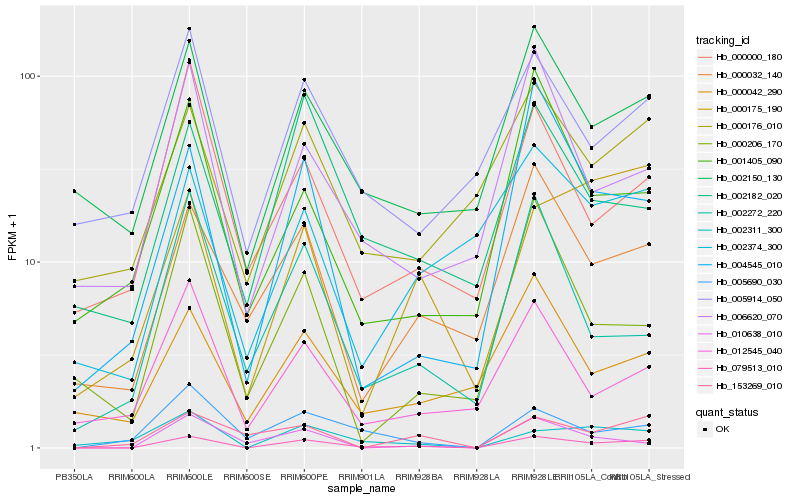
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079513_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_004545_010 |
0.2247873691 |
- |
- |
acyl carrier protein [Ricinus communis] |
| 3 |
Hb_000032_140 |
0.2376777501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002150_130 |
0.2377878905 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_002311_300 |
0.2477152919 |
- |
- |
PREDICTED: J protein JJJ2 [Jatropha curcas] |
| 6 |
Hb_005690_030 |
0.2531432357 |
- |
- |
hypothetical protein VITISV_020378 [Vitis vinifera] |
| 7 |
Hb_006620_070 |
0.2546823573 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 8 |
Hb_000176_010 |
0.255192722 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000175_190 |
0.2555028969 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 10 |
Hb_002272_220 |
0.2577355876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002374_300 |
0.2581673753 |
- |
- |
hypothetical protein JCGZ_12140 [Jatropha curcas] |
| 12 |
Hb_002182_020 |
0.2583524338 |
- |
- |
Thylakoid lumenal 17.4 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_010638_010 |
0.2590083092 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 14 |
Hb_153269_010 |
0.2591901347 |
- |
- |
hypothetical protein CICLE_v10029937mg [Citrus clementina] |
| 15 |
Hb_012545_040 |
0.2594603843 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_005914_050 |
0.259815437 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000042_290 |
0.262692729 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000000_180 |
0.2630718569 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_000206_170 |
0.2644470197 |
- |
- |
hypothetical protein JCGZ_25514 [Jatropha curcas] |
| 20 |
Hb_001405_090 |
0.2645615918 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 5, chloroplastic [Jatropha curcas] |