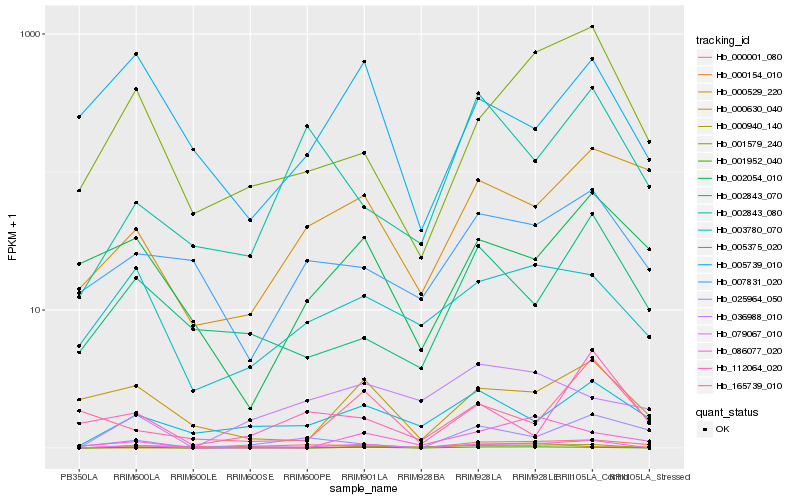
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079067_010 |
0.0 |
- |
- |
PREDICTED: protein SPT2 homolog [Malus domestica] |
| 2 |
Hb_001952_040 |
0.281998839 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_001579_240 |
0.2983113944 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 4 |
Hb_000001_080 |
0.3115094166 |
- |
- |
- |
| 5 |
Hb_005375_020 |
0.33133676 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 1-like [Nicotiana sylvestris] |
| 6 |
Hb_003780_070 |
0.3433586684 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 7 |
Hb_000630_040 |
0.3449821458 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 8 |
Hb_002843_070 |
0.3550832405 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_007831_020 |
0.3707995651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002843_080 |
0.3737643281 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 11 |
Hb_005739_010 |
0.3744317028 |
- |
- |
hypothetical protein JCGZ_04016 [Jatropha curcas] |
| 12 |
Hb_000529_220 |
0.3754643327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_025964_050 |
0.3762206494 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 14 |
Hb_165739_010 |
0.3770470471 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_002054_010 |
0.3770985949 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 16 |
Hb_086077_020 |
0.3776763323 |
- |
- |
hypothetical protein PHAVU_001G054900g [Phaseolus vulgaris] |
| 17 |
Hb_112064_020 |
0.3779955724 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 18 |
Hb_036988_010 |
0.3783021388 |
- |
- |
hypothetical protein JCGZ_25496 [Jatropha curcas] |
| 19 |
Hb_000940_140 |
0.380707445 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 20 |
Hb_000154_010 |
0.3820102921 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |