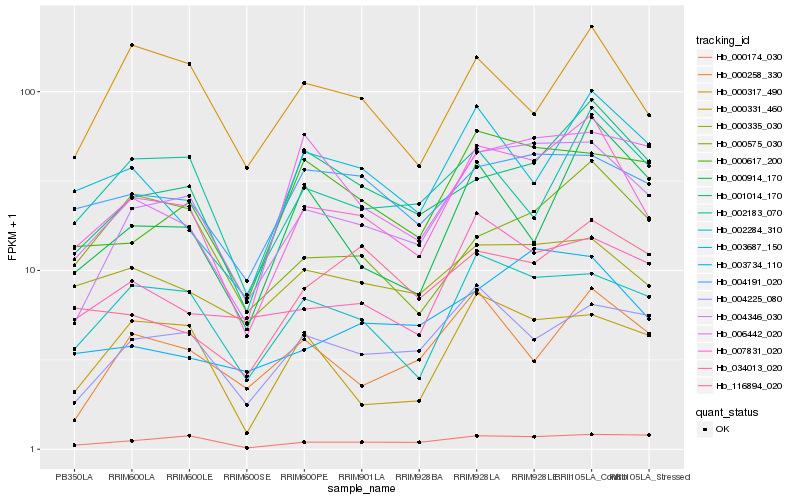
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007831_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004346_030 |
0.1207603505 |
- |
- |
sucrose phosphate phosphatase, putative [Ricinus communis] |
| 3 |
Hb_002284_310 |
0.1481141557 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001014_170 |
0.1511833899 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_003687_150 |
0.1519572045 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000335_030 |
0.1618162445 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17033 [Jatropha curcas] |
| 7 |
Hb_034013_020 |
0.1619684704 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 8 |
Hb_002183_070 |
0.167761752 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_004191_020 |
0.168435146 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 10 |
Hb_000575_030 |
0.1700145003 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_003734_110 |
0.1706343202 |
- |
- |
PREDICTED: cytochrome c-type biogenesis ccda-like chloroplastic protein [Jatropha curcas] |
| 12 |
Hb_000617_200 |
0.1743056995 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 13 |
Hb_004225_080 |
0.1746647115 |
- |
- |
- |
| 14 |
Hb_000914_170 |
0.1747842326 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 15 |
Hb_000174_030 |
0.1748375714 |
- |
- |
hypothetical protein RCOM_1077370 [Ricinus communis] |
| 16 |
Hb_000317_490 |
0.1750943299 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 17 |
Hb_000331_460 |
0.1809313681 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 18 |
Hb_116894_020 |
0.1816790796 |
- |
- |
hypothetical protein POPTR_0008s07660g [Populus trichocarpa] |
| 19 |
Hb_000258_330 |
0.1826731622 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 20 |
Hb_006442_020 |
0.1834524835 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |