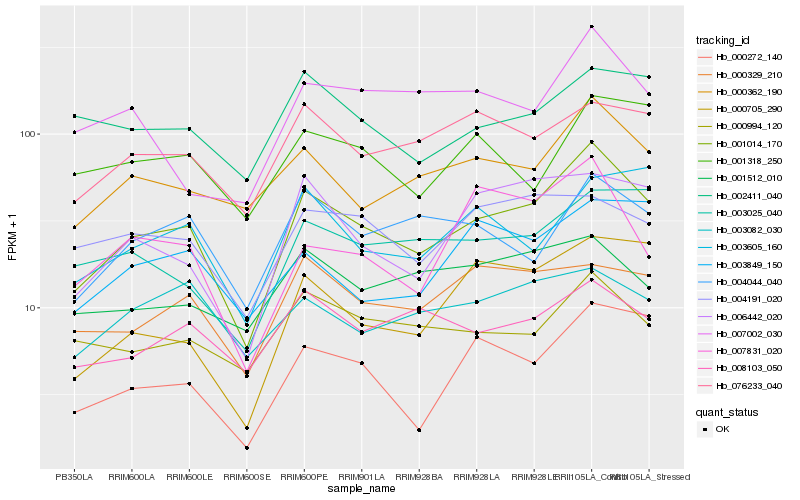
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001014_170 |
0.0 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_000362_190 |
0.1226432243 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 3 |
Hb_000272_140 |
0.1328586755 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 4 |
Hb_006442_020 |
0.1358661081 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 5 |
Hb_004044_040 |
0.137979711 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 6 |
Hb_076233_040 |
0.1400988311 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000994_120 |
0.1427046327 |
- |
- |
unknown [Populus trichocarpa] |
| 8 |
Hb_003025_040 |
0.1435611416 |
- |
- |
PREDICTED: elongin-A [Jatropha curcas] |
| 9 |
Hb_000329_210 |
0.1449417367 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 10 |
Hb_008103_050 |
0.1449600662 |
- |
- |
hypothetical protein JCGZ_07422 [Jatropha curcas] |
| 11 |
Hb_000705_290 |
0.1453778811 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |
| 12 |
Hb_003605_160 |
0.1490045856 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 13 |
Hb_007831_020 |
0.1511833899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004191_020 |
0.1514383949 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 15 |
Hb_003082_030 |
0.1521580442 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003849_150 |
0.1526181238 |
- |
- |
PREDICTED: uncharacterized protein LOC105644698 [Jatropha curcas] |
| 17 |
Hb_001512_010 |
0.1526216305 |
- |
- |
PREDICTED: uncharacterized protein LOC105633759 [Jatropha curcas] |
| 18 |
Hb_001318_250 |
0.1528952921 |
- |
- |
PREDICTED: uncharacterized protein LOC105642666 [Jatropha curcas] |
| 19 |
Hb_007002_030 |
0.1529002012 |
- |
- |
PREDICTED: transmembrane protein 205 [Jatropha curcas] |
| 20 |
Hb_002411_040 |
0.1532561166 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 6 [Jatropha curcas] |