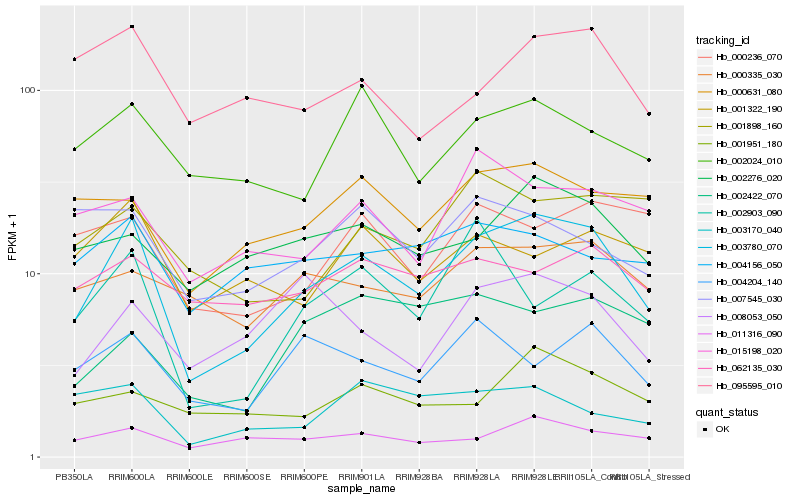
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003780_070 |
0.0 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 2 |
Hb_011316_090 |
0.1729026263 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 3 |
Hb_002276_020 |
0.1758038423 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000631_080 |
0.1803983243 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Jatropha curcas] |
| 5 |
Hb_002024_010 |
0.1843102192 |
- |
- |
hypothetical protein B456_003G0564002, partial [Gossypium raimondii] |
| 6 |
Hb_015198_020 |
0.1857481608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000335_030 |
0.1863097646 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17033 [Jatropha curcas] |
| 8 |
Hb_004204_140 |
0.1867703506 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1470470 [Ricinus communis] |
| 9 |
Hb_002903_090 |
0.1878127402 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 10 |
Hb_001951_180 |
0.1905371241 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_004156_050 |
0.1923830358 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 12 |
Hb_000236_070 |
0.1933507158 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_095595_010 |
0.1938491554 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 14 |
Hb_002422_070 |
0.1941045448 |
- |
- |
PREDICTED: putative protein TPRXL [Malus domestica] |
| 15 |
Hb_001322_190 |
0.1941715742 |
- |
- |
PREDICTED: uncharacterized protein LOC105632381 [Jatropha curcas] |
| 16 |
Hb_008053_050 |
0.1955523493 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001898_160 |
0.195600526 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 18 |
Hb_062135_030 |
0.1971395052 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 19 |
Hb_003170_040 |
0.198382317 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 20 |
Hb_007545_030 |
0.1987244617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |