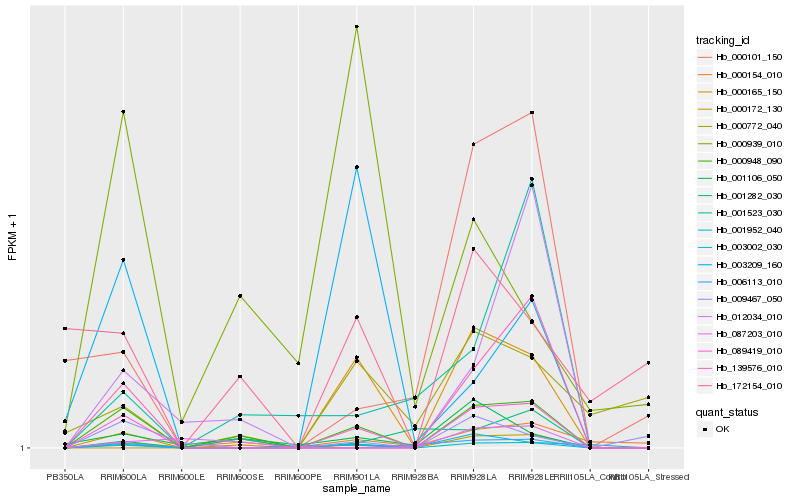
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000948_090 |
0.0 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 2 |
Hb_089419_010 |
0.1922724907 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 3 |
Hb_001106_050 |
0.3392587345 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_001952_040 |
0.3494898449 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_000165_150 |
0.3512620663 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 6 |
Hb_139576_010 |
0.3625068408 |
- |
- |
PREDICTED: uncharacterized protein LOC105795953 [Gossypium raimondii] |
| 7 |
Hb_000154_010 |
0.3680432246 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_000172_130 |
0.3767514568 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 9 |
Hb_001523_030 |
0.3877938526 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_003002_030 |
0.3904261574 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 11 |
Hb_000101_150 |
0.399095464 |
- |
- |
- |
| 12 |
Hb_012034_010 |
0.3993008182 |
- |
- |
- |
| 13 |
Hb_009467_050 |
0.399504921 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 14 |
Hb_001282_030 |
0.4066403625 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 15 |
Hb_172154_010 |
0.4085424324 |
- |
- |
hypothetical protein JCGZ_25247 [Jatropha curcas] |
| 16 |
Hb_000939_010 |
0.4100212796 |
- |
- |
PREDICTED: transcription factor bHLH113-like [Jatropha curcas] |
| 17 |
Hb_087203_010 |
0.4106090108 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 68-like [Pyrus x bretschneideri] |
| 18 |
Hb_003209_160 |
0.4108402498 |
- |
- |
PREDICTED: putative SNAP25 homologous protein SNAP30 [Populus euphratica] |
| 19 |
Hb_000772_040 |
0.4110205223 |
- |
- |
PREDICTED: uncharacterized protein LOC105036273 [Elaeis guineensis] |
| 20 |
Hb_006113_010 |
0.4178476359 |
- |
- |
hypothetical protein VITISV_015014 [Vitis vinifera] |