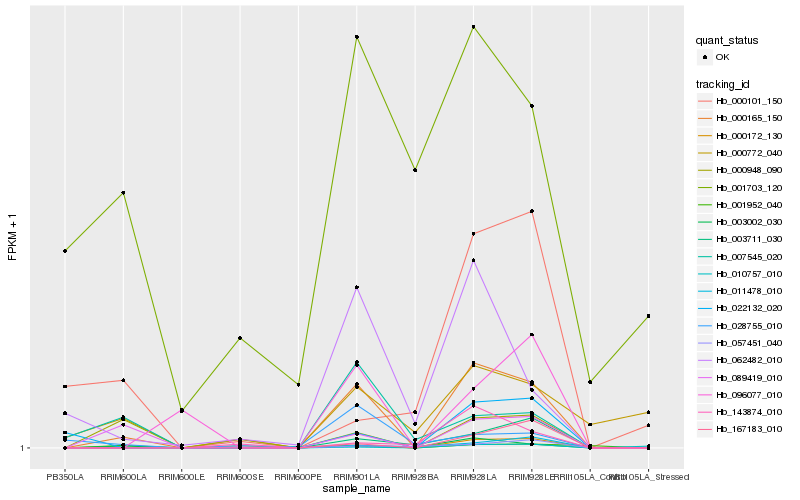
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000165_150 |
0.0 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 2 |
Hb_089419_010 |
0.2769666907 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 3 |
Hb_003711_030 |
0.3168335253 |
- |
- |
PREDICTED: uncharacterized protein LOC102621950 [Citrus sinensis] |
| 4 |
Hb_062482_010 |
0.3182766586 |
- |
- |
- |
| 5 |
Hb_000172_130 |
0.3216594804 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 6 |
Hb_011478_010 |
0.3304438644 |
- |
- |
- |
| 7 |
Hb_167183_010 |
0.3323340639 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 8 |
Hb_003002_030 |
0.3352792699 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 9 |
Hb_096077_010 |
0.3355895818 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 10 |
Hb_022132_020 |
0.3357130994 |
- |
- |
hypothetical protein TRIUR3_12476 [Triticum urartu] |
| 11 |
Hb_000948_090 |
0.3512620663 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 12 |
Hb_007545_020 |
0.3599612311 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Nicotiana tomentosiformis] |
| 13 |
Hb_001952_040 |
0.3601124752 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_143874_010 |
0.3724095985 |
- |
- |
PREDICTED: uncharacterized protein LOC105794100 [Gossypium raimondii] |
| 15 |
Hb_010757_010 |
0.3901907368 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 16 |
Hb_028755_010 |
0.3957402638 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_057451_040 |
0.3974352944 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_000772_040 |
0.3981242649 |
- |
- |
PREDICTED: uncharacterized protein LOC105036273 [Elaeis guineensis] |
| 19 |
Hb_000101_150 |
0.406931857 |
- |
- |
- |
| 20 |
Hb_001703_120 |
0.4087990205 |
- |
- |
PREDICTED: uncharacterized protein LOC105116966 isoform X2 [Populus euphratica] |