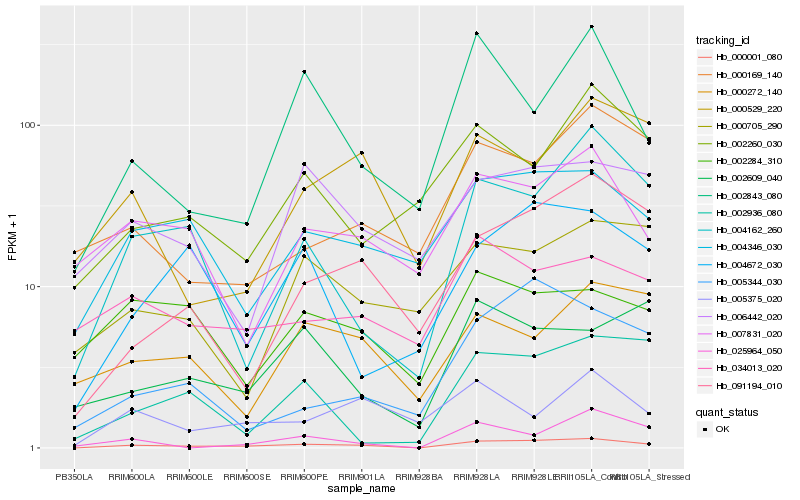
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000001_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_004346_030 |
0.2094629428 |
- |
- |
sucrose phosphate phosphatase, putative [Ricinus communis] |
| 3 |
Hb_004162_260 |
0.2221873499 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_091194_010 |
0.2266124226 |
- |
- |
hypothetical protein POPTR_0006s05430g [Populus trichocarpa] |
| 5 |
Hb_004672_030 |
0.2333970676 |
- |
- |
proline oxidase [Manihot esculenta] |
| 6 |
Hb_007831_020 |
0.2385935864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002843_080 |
0.2441463695 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 8 |
Hb_034013_020 |
0.2443265296 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 9 |
Hb_025964_050 |
0.2448907906 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 10 |
Hb_002936_080 |
0.2454104465 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 11 |
Hb_006442_020 |
0.245730707 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 12 |
Hb_000529_220 |
0.2479269887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002609_040 |
0.2480237787 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 14 |
Hb_005344_030 |
0.249092185 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Jatropha curcas] |
| 15 |
Hb_005375_020 |
0.2523396675 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 1-like [Nicotiana sylvestris] |
| 16 |
Hb_002260_030 |
0.2531790183 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 17 |
Hb_000272_140 |
0.2538151926 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 18 |
Hb_002284_310 |
0.2540229942 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000169_140 |
0.2543130921 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 20 |
Hb_000705_290 |
0.2545738027 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |