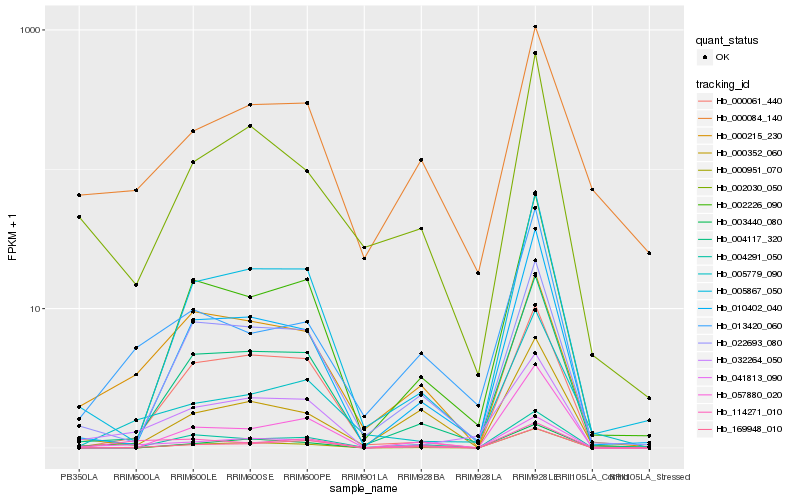
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041813_090 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_J01097 [Eucalyptus grandis] |
| 2 |
Hb_032264_050 |
0.1512025315 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 3 |
Hb_004117_320 |
0.1567922798 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 4 |
Hb_002030_050 |
0.1662106386 |
- |
- |
classical arabinogalactan protein 7 precursor [Jatropha curcas] |
| 5 |
Hb_169948_010 |
0.1743541242 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |
| 6 |
Hb_013420_060 |
0.1771798502 |
- |
- |
NADH dehydrogenase subunit 7 [Nymphaea sp. Bergthorsson 0401] |
| 7 |
Hb_010402_040 |
0.1778188144 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 8 |
Hb_005867_050 |
0.1778968494 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 9 |
Hb_005779_090 |
0.1864014891 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 10 |
Hb_000951_070 |
0.1901217824 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 11 |
Hb_022693_080 |
0.1947463449 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 12 |
Hb_000061_440 |
0.195084887 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000215_230 |
0.1980825112 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 14 |
Hb_003440_080 |
0.2030280876 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 15 |
Hb_004291_050 |
0.2050727121 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 16 |
Hb_114271_010 |
0.2053149156 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 17 |
Hb_000084_140 |
0.2053722727 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 18 |
Hb_002226_090 |
0.2069780655 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 19 |
Hb_057880_020 |
0.2080962485 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 20 |
Hb_000352_060 |
0.2088401573 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |