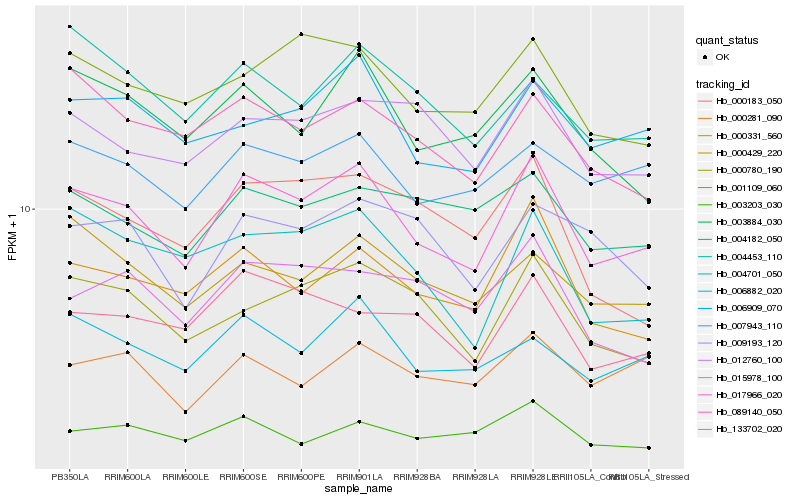
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017966_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03560, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000331_560 |
0.0860208523 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 3 |
Hb_006882_020 |
0.0899670148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006909_070 |
0.0901052219 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_000780_190 |
0.0905581376 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 6 |
Hb_001109_060 |
0.0913781212 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_003884_030 |
0.0927514609 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 8 |
Hb_007943_110 |
0.0950214065 |
- |
- |
PREDICTED: basic salivary proline-rich protein 3 [Jatropha curcas] |
| 9 |
Hb_004701_050 |
0.0950526666 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 10 |
Hb_133702_020 |
0.0965982158 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_012760_100 |
0.0971821632 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 12 |
Hb_089140_050 |
0.0981673647 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 13 |
Hb_000429_220 |
0.0983020391 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004182_050 |
0.1029017753 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_015978_100 |
0.1033598032 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH6 [Jatropha curcas] |
| 16 |
Hb_000281_090 |
0.1046495971 |
transcription factor |
TF Family: SNF2 |
hypothetical protein PRUPE_ppa000063mg [Prunus persica] |
| 17 |
Hb_009193_120 |
0.1052849311 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 18 |
Hb_004453_110 |
0.1062923039 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 19 |
Hb_003203_030 |
0.1068543048 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 20 |
Hb_000183_050 |
0.1070008093 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |