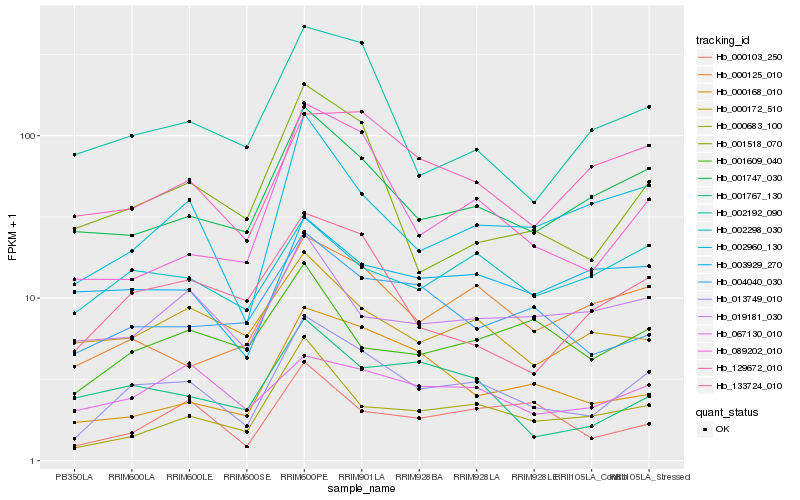
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013749_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001747_030 |
0.1482113775 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 3 |
Hb_002960_130 |
0.156723001 |
- |
- |
hypothetical protein B456_008G036100 [Gossypium raimondii] |
| 4 |
Hb_000683_100 |
0.1578754051 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 5 |
Hb_000125_010 |
0.1662225862 |
transcription factor |
TF Family: mTERF |
- |
| 6 |
Hb_129672_010 |
0.1672948576 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 7 |
Hb_089202_010 |
0.1674682607 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 8 |
Hb_133724_010 |
0.1689264874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000103_250 |
0.1698785499 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 4B isoform X3 [Jatropha curcas] |
| 10 |
Hb_002298_030 |
0.1719391382 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 11 |
Hb_004040_030 |
0.1721410254 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002192_090 |
0.1730571881 |
- |
- |
hypothetical protein AMTR_s00029p00055000 [Amborella trichopoda] |
| 13 |
Hb_067130_010 |
0.1736932355 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_001767_130 |
0.1779491607 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 15 |
Hb_000172_510 |
0.1779869525 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |
| 16 |
Hb_019181_030 |
0.17838262 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 17 |
Hb_000168_010 |
0.180915626 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3-like isoform X1 [Populus euphratica] |
| 18 |
Hb_001518_070 |
0.1811473597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003929_270 |
0.1833712071 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_001609_040 |
0.1836525623 |
- |
- |
hypothetical protein POPTR_0017s05650g [Populus trichocarpa] |