| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
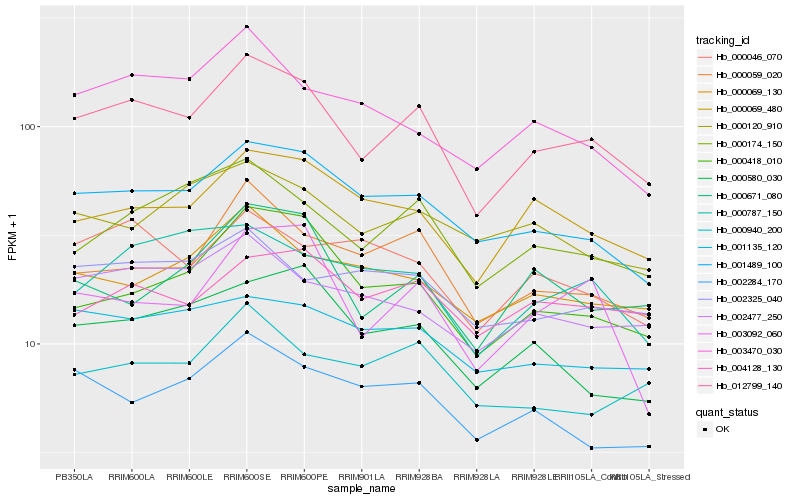
Hb_012799_140 |
0.0 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 2 |
Hb_001489_100 |
0.0852712812 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 3 |
Hb_000174_150 |
0.0871301931 |
- |
- |
stearoyl-acyl-carrier protein desaturase [Ricinus communis] |
| 4 |
Hb_000059_020 |
0.0919951781 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 5 |
Hb_000069_480 |
0.0949202726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000418_010 |
0.0966694812 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 7 |
Hb_002477_250 |
0.0978931109 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 8 |
Hb_000069_130 |
0.1035754013 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000787_150 |
0.1036847317 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 10 |
Hb_004128_130 |
0.1043361942 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001135_120 |
0.1049870059 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000671_080 |
0.1055531547 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 13 |
Hb_003470_030 |
0.1057236161 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 14 |
Hb_003092_060 |
0.1058255791 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 15 |
Hb_000580_030 |
0.1064557328 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 16 |
Hb_002284_170 |
0.1075079106 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 17 |
Hb_002325_040 |
0.1086252157 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000046_070 |
0.108825007 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 19 |
Hb_000940_200 |
0.1094717901 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 20 |
Hb_000120_910 |
0.1102063939 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |