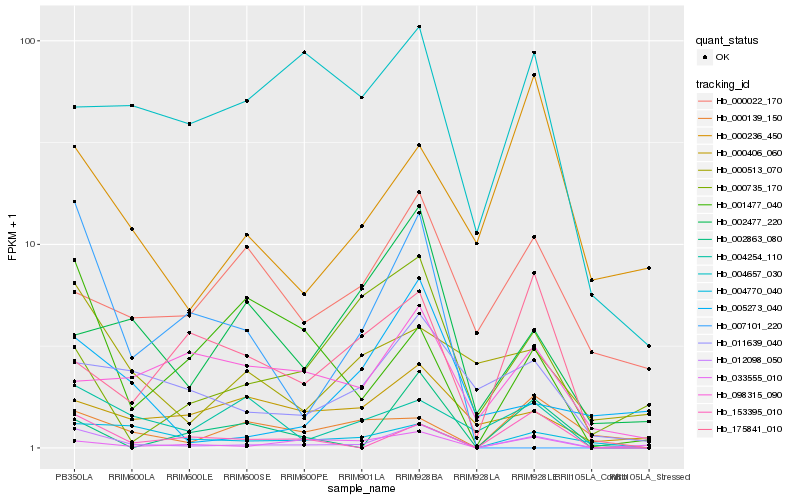
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012098_050 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |
| 2 |
Hb_004770_040 |
0.2727253261 |
- |
- |
PREDICTED: serine carboxypeptidase-like 31 [Eucalyptus grandis] |
| 3 |
Hb_153395_010 |
0.2734937636 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_011639_040 |
0.2957578365 |
- |
- |
hypothetical protein M569_08265, partial [Genlisea aurea] |
| 5 |
Hb_002863_080 |
0.3056688218 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 6 |
Hb_033555_010 |
0.3074826765 |
- |
- |
- |
| 7 |
Hb_004254_110 |
0.318475415 |
- |
- |
- |
| 8 |
Hb_005273_040 |
0.3230266378 |
- |
- |
- |
| 9 |
Hb_000513_070 |
0.324863936 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 42 [Jatropha curcas] |
| 10 |
Hb_002477_220 |
0.3307261709 |
- |
- |
hypothetical protein POPTR_0017s06650g [Populus trichocarpa] |
| 11 |
Hb_000139_150 |
0.3354412785 |
- |
- |
transposon protein, putative, Mutator sub-class [Oryza sativa Japonica Group] |
| 12 |
Hb_175841_010 |
0.3377277081 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 13 |
Hb_000406_060 |
0.3395910038 |
- |
- |
hypothetical protein CICLE_v10000115mg [Citrus clementina] |
| 14 |
Hb_007101_220 |
0.3413231662 |
- |
- |
Gibberellin-regulated protein 3 precursor, putative [Ricinus communis] |
| 15 |
Hb_098315_090 |
0.3428688347 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 16 |
Hb_004657_030 |
0.3432873799 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 17 |
Hb_000022_170 |
0.3441644006 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 18 |
Hb_000236_450 |
0.3444641573 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 19 |
Hb_000735_170 |
0.3460577206 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |
| 20 |
Hb_001477_040 |
0.3476678054 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |