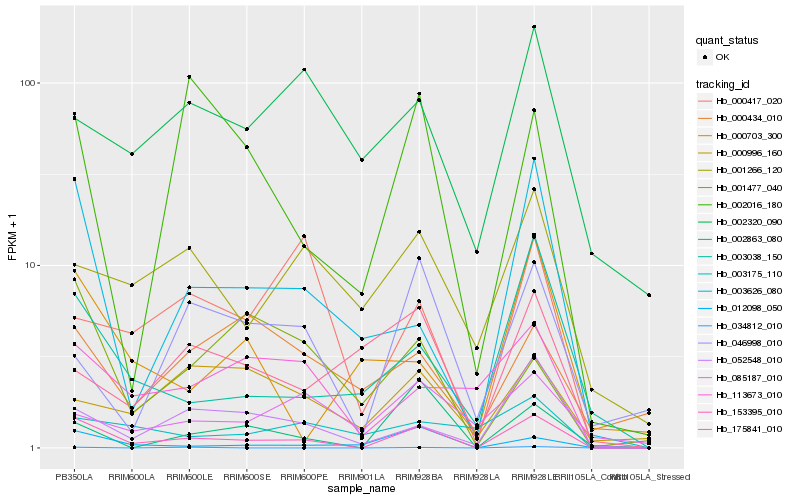
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_153395_010 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003626_080 |
0.2608123124 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_052548_010 |
0.2683987904 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 4 |
Hb_012098_050 |
0.2734937636 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 isoform X1 [Nicotiana tomentosiformis] |
| 5 |
Hb_003038_150 |
0.2800546704 |
- |
- |
- |
| 6 |
Hb_002863_080 |
0.3007810866 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 7 |
Hb_113673_010 |
0.3027264304 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 8 |
Hb_002016_180 |
0.3079369375 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 9 |
Hb_001266_120 |
0.3144538788 |
- |
- |
auxilin, putative [Ricinus communis] |
| 10 |
Hb_001477_040 |
0.321652643 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 11 |
Hb_046998_010 |
0.3218076447 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 12 |
Hb_000703_300 |
0.3309849702 |
- |
- |
- |
| 13 |
Hb_002320_090 |
0.3313010816 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 14 |
Hb_000417_020 |
0.3322095043 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 15 |
Hb_003175_110 |
0.3332639385 |
- |
- |
- |
| 16 |
Hb_175841_010 |
0.3347953542 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 17 |
Hb_000996_160 |
0.3365660097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000434_010 |
0.3394289913 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 19 |
Hb_034812_010 |
0.3413414471 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_085187_010 |
0.3420068935 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |