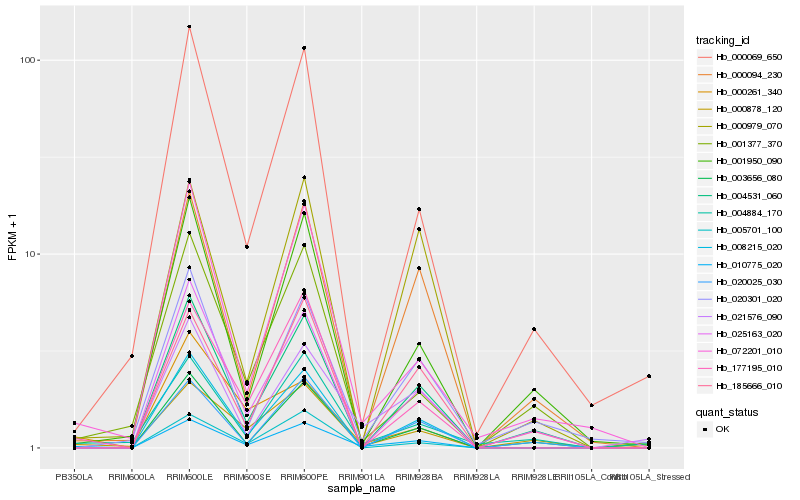
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010775_020 |
0.0 |
transcription factor |
TF Family: ERF |
Floral homeotic protein APETALA2, putative [Ricinus communis] |
| 2 |
Hb_008215_020 |
0.1469510822 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_025163_020 |
0.1565790531 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 4 |
Hb_072201_010 |
0.1653575676 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 5 |
Hb_003656_080 |
0.1680120988 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 6 |
Hb_185666_010 |
0.1688243188 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 7 |
Hb_177195_010 |
0.1692179443 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 3, partial [Theobroma cacao] |
| 8 |
Hb_005701_100 |
0.1718390892 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 9 |
Hb_000878_120 |
0.1733439185 |
- |
- |
PREDICTED: protein downstream neighbor of Son [Jatropha curcas] |
| 10 |
Hb_000094_230 |
0.1791174647 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 11 |
Hb_004531_060 |
0.1792592515 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 12 |
Hb_000069_650 |
0.1854392944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004884_170 |
0.186593312 |
- |
- |
hypothetical protein AALP_AA1G067000 [Arabis alpina] |
| 14 |
Hb_000979_070 |
0.192224728 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 15 |
Hb_020025_030 |
0.1945910452 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 16 |
Hb_000261_340 |
0.1946048318 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 17 |
Hb_001950_090 |
0.195537958 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 18 |
Hb_021576_090 |
0.1968828263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001377_370 |
0.2007102546 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 20 |
Hb_020301_020 |
0.2023380115 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |