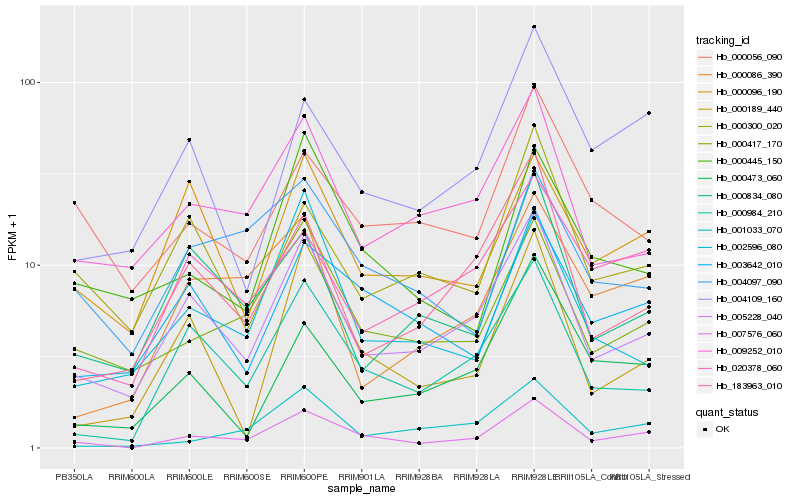
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007576_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_005228_040 |
0.1430616494 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 3 |
Hb_000417_170 |
0.1587896694 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |
| 4 |
Hb_000984_210 |
0.1608041964 |
- |
- |
PREDICTED: uncharacterized protein LOC104451821 [Eucalyptus grandis] |
| 5 |
Hb_003642_010 |
0.1849907962 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_001033_070 |
0.1857170262 |
- |
- |
seryl-tRNA synthetase, putative [Ricinus communis] |
| 7 |
Hb_009252_010 |
0.1893365492 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 8 |
Hb_004109_160 |
0.1976210575 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000189_440 |
0.1978558094 |
- |
- |
hypothetical protein JCGZ_23470 [Jatropha curcas] |
| 10 |
Hb_183963_010 |
0.1978840537 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 11 |
Hb_000445_150 |
0.1980125201 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 12 |
Hb_004097_090 |
0.2001605346 |
- |
- |
PREDICTED: uncharacterized protein LOC105634517 [Jatropha curcas] |
| 13 |
Hb_020378_060 |
0.2013746688 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 14 |
Hb_000086_390 |
0.2021362893 |
- |
- |
PREDICTED: uncharacterized protein LOC105644503 [Jatropha curcas] |
| 15 |
Hb_000096_190 |
0.2044589508 |
- |
- |
unknown [Lotus japonicus] |
| 16 |
Hb_002596_080 |
0.2056623462 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 17 |
Hb_000834_080 |
0.208395913 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 18 |
Hb_000300_020 |
0.2088470671 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000473_060 |
0.209584774 |
- |
- |
PREDICTED: psbP domain-containing protein 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000056_090 |
0.2100916811 |
- |
- |
UDP-galactose transporter 3 isoform 1 [Theobroma cacao] |