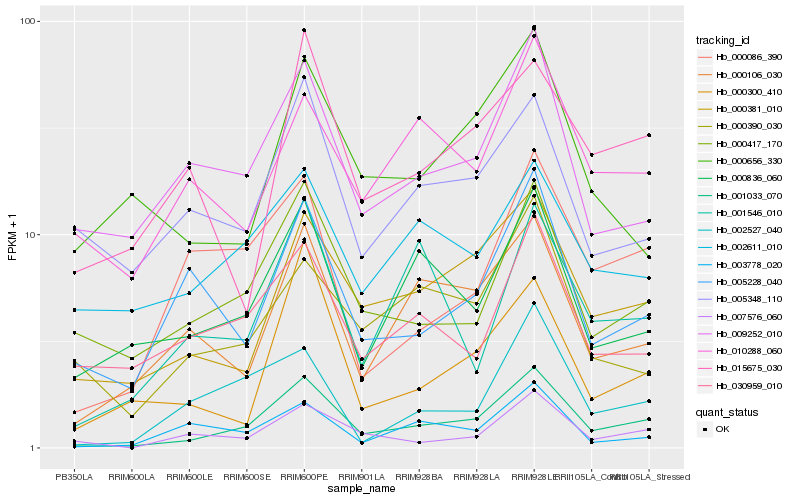
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001033_070 |
0.0 |
- |
- |
seryl-tRNA synthetase, putative [Ricinus communis] |
| 2 |
Hb_000381_010 |
0.1571615532 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000417_170 |
0.1763504353 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |
| 4 |
Hb_000106_030 |
0.179189276 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_000836_060 |
0.1849834821 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 6 |
Hb_007576_060 |
0.1857170262 |
- |
- |
- |
| 7 |
Hb_002527_040 |
0.1909672949 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 8 |
Hb_000086_390 |
0.1912123037 |
- |
- |
PREDICTED: uncharacterized protein LOC105644503 [Jatropha curcas] |
| 9 |
Hb_009252_010 |
0.1922961017 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 10 |
Hb_005228_040 |
0.1956952262 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 11 |
Hb_000300_410 |
0.1966361127 |
- |
- |
hypothetical protein POPTR_0013s05860g [Populus trichocarpa] |
| 12 |
Hb_030959_010 |
0.2033649777 |
- |
- |
cullin, putative [Ricinus communis] |
| 13 |
Hb_001546_010 |
0.2038167927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000390_030 |
0.2053083291 |
- |
- |
hypothetical protein ZEAMMB73_364791 [Zea mays] |
| 15 |
Hb_003778_020 |
0.2056900377 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 16 |
Hb_015675_030 |
0.2073666467 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002611_010 |
0.2077896741 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_010288_060 |
0.2081236741 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 19 |
Hb_005348_110 |
0.2096238474 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 20 |
Hb_000656_330 |
0.2103108636 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |