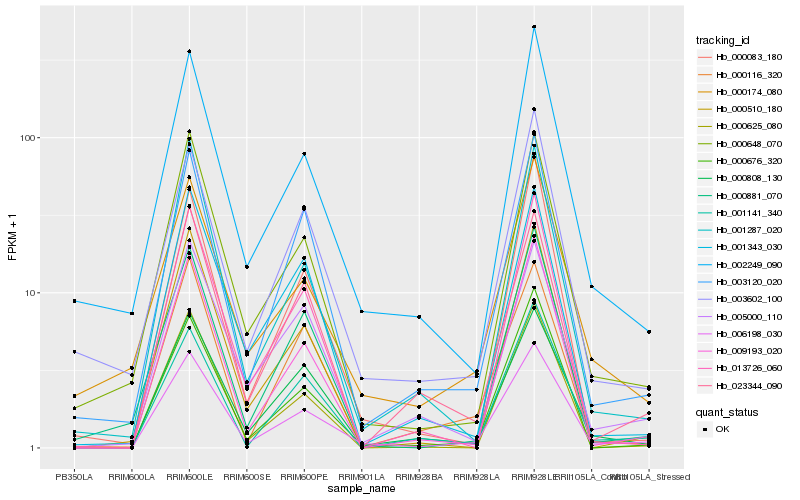
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006198_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003120_020 |
0.1065400505 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 13-like [Prunus mume] |
| 3 |
Hb_009193_020 |
0.1086306638 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 4 |
Hb_000808_130 |
0.1091550726 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |
| 5 |
Hb_000676_320 |
0.1102321435 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 6 |
Hb_000648_070 |
0.1113023208 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 7 |
Hb_002249_090 |
0.1177338251 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001141_340 |
0.1220836652 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 9 |
Hb_001287_020 |
0.122407191 |
- |
- |
SulA [Manihot esculenta] |
| 10 |
Hb_023344_090 |
0.1242472731 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 11 |
Hb_000116_320 |
0.1293572773 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 12 |
Hb_003602_100 |
0.1295384949 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000510_180 |
0.1298567268 |
- |
- |
unknown [Lotus japonicus] |
| 14 |
Hb_013726_060 |
0.1303702118 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 15 |
Hb_000625_080 |
0.1313471463 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005000_110 |
0.131387664 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000174_080 |
0.1314592557 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |
| 18 |
Hb_000881_070 |
0.1315204228 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000083_180 |
0.132739577 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 20 |
Hb_001343_030 |
0.136566605 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |