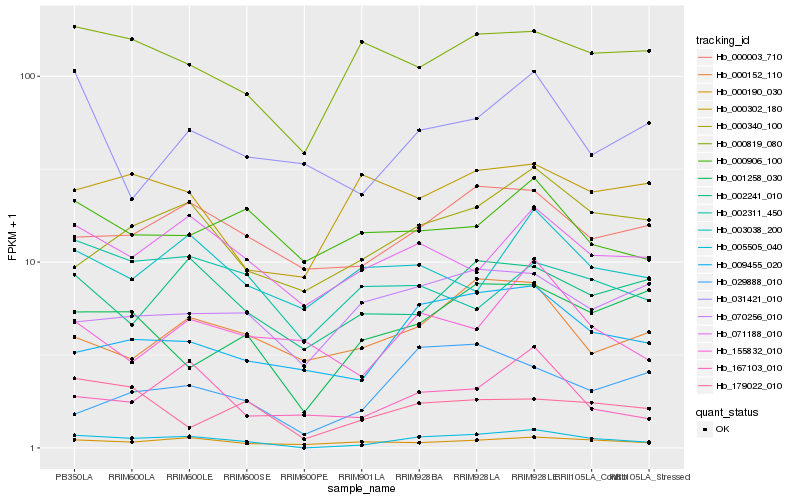
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005505_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_009455_020 |
0.1935645216 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 3 |
Hb_000340_100 |
0.201185297 |
- |
- |
PREDICTED: protein PALE CRESS, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000003_710 |
0.2021330379 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 5 |
Hb_071188_010 |
0.2024745985 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 6 |
Hb_000190_030 |
0.2047993258 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA-like [Pyrus x bretschneideri] |
| 7 |
Hb_167103_010 |
0.206977671 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 8 |
Hb_002241_010 |
0.2101702057 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 9 |
Hb_001258_030 |
0.2103227522 |
- |
- |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 10 |
Hb_000819_080 |
0.2148142797 |
- |
- |
Plastid-lipid-associated protein, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000152_110 |
0.2203869006 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_179022_010 |
0.2205230083 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 13 |
Hb_003038_200 |
0.222516677 |
desease resistance |
Gene Name: MMR_HSR1 |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |
| 14 |
Hb_155832_010 |
0.223275072 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 15 |
Hb_029888_010 |
0.2270370152 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Nicotiana tomentosiformis] |
| 16 |
Hb_070256_010 |
0.2274874975 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105630457 [Jatropha curcas] |
| 17 |
Hb_002311_450 |
0.2290853221 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 18 |
Hb_031421_010 |
0.2297273455 |
- |
- |
PREDICTED: copper chaperone for superoxide dismutase, chloroplastic/cytosolic [Jatropha curcas] |
| 19 |
Hb_000302_180 |
0.23025844 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000906_100 |
0.2302731062 |
- |
- |
PREDICTED: uncharacterized protein LOC105648070 [Jatropha curcas] |