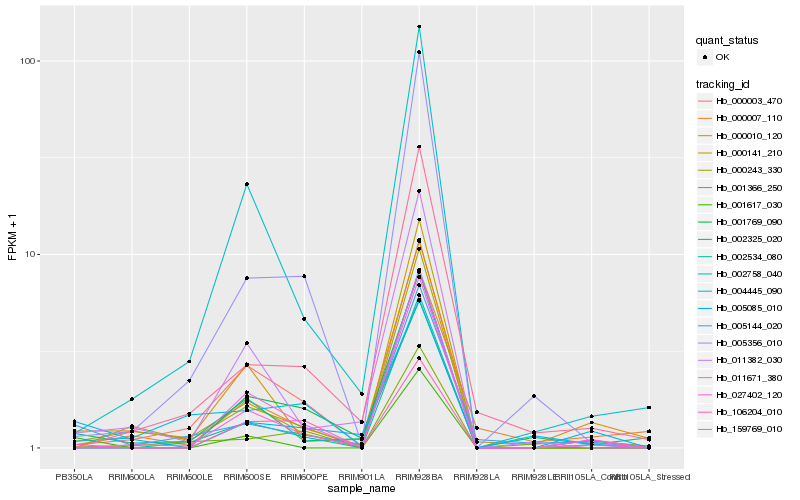
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005085_010 |
0.0 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 2 |
Hb_005144_020 |
0.1983499087 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0009s01270g [Populus trichocarpa] |
| 3 |
Hb_000010_120 |
0.2163194647 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 4 |
Hb_001769_090 |
0.2251525818 |
- |
- |
PREDICTED: uncharacterized protein LOC105630842 [Jatropha curcas] |
| 5 |
Hb_011382_030 |
0.2265449545 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 6 |
Hb_002325_020 |
0.2343709746 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_027402_120 |
0.2409974884 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_159769_010 |
0.2494198131 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 9 |
Hb_011671_380 |
0.2507588706 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 10 |
Hb_106204_010 |
0.2522922601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001617_030 |
0.2539230133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001366_250 |
0.2555186655 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 4-like [Jatropha curcas] |
| 13 |
Hb_000141_210 |
0.2566368576 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Camelina sativa] |
| 14 |
Hb_000007_110 |
0.2586076839 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 15 |
Hb_002758_040 |
0.2587962443 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000243_330 |
0.2637268241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004445_090 |
0.2646720711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002534_080 |
0.2648151063 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000003_470 |
0.264942156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005356_010 |
0.2654200099 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |