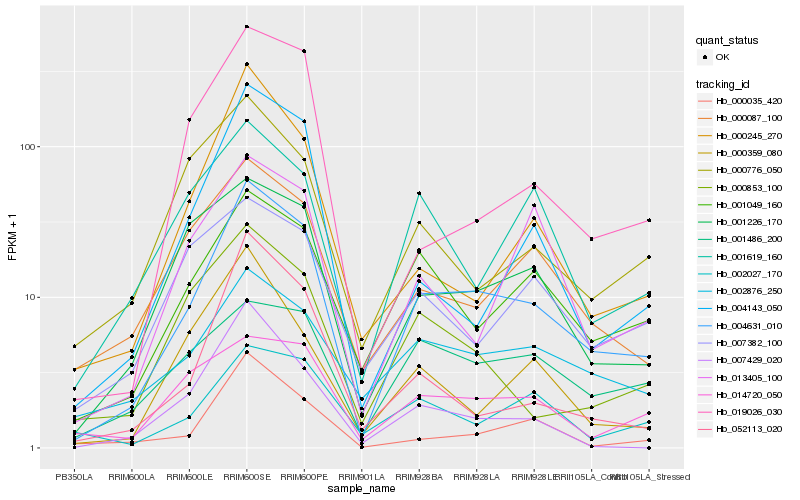
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004631_010 |
0.0 |
- |
- |
Early nodulin 20 precursor, putative [Ricinus communis] |
| 2 |
Hb_001049_160 |
0.1715059471 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 3 |
Hb_001226_170 |
0.1802468398 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 4 |
Hb_000087_100 |
0.1812820276 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 5 |
Hb_002876_250 |
0.1850319447 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 6 |
Hb_000035_420 |
0.187221337 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 7 |
Hb_004143_050 |
0.1997084147 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 8 |
Hb_014720_050 |
0.1997657728 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 9 |
Hb_000245_270 |
0.1999992598 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 10 |
Hb_000853_100 |
0.2006849737 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 11 |
Hb_007429_020 |
0.2020471874 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_002027_170 |
0.2027159593 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 13 |
Hb_052113_020 |
0.2033713806 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 14 |
Hb_019026_030 |
0.2051164382 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 15 |
Hb_001486_200 |
0.2071363346 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 16 |
Hb_001619_160 |
0.207190821 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 17 |
Hb_013405_100 |
0.2087641904 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 18 |
Hb_000776_050 |
0.2097806305 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 19 |
Hb_007382_100 |
0.2117341784 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000359_080 |
0.212308967 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |