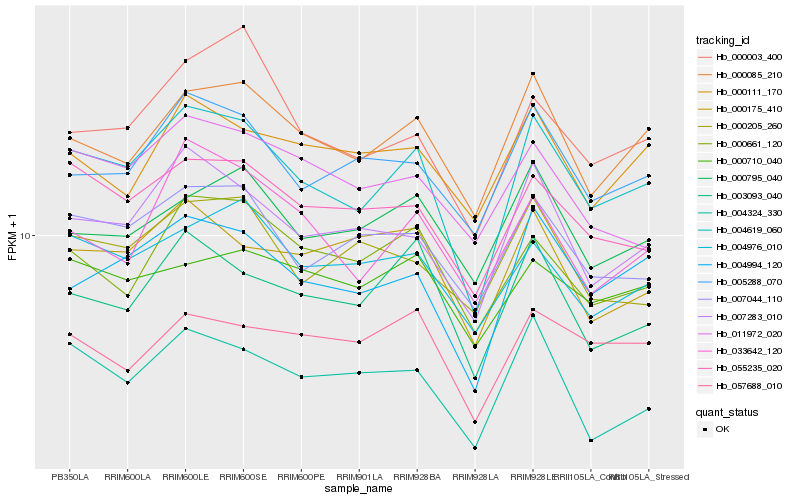
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004619_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 2 |
Hb_004994_120 |
0.0667806391 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 13-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_007283_010 |
0.0737537258 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 4 |
Hb_000085_210 |
0.0761134994 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 5 |
Hb_055235_020 |
0.0813551788 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007044_110 |
0.0813610437 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_003093_040 |
0.0838595114 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_005288_070 |
0.085230533 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_004976_010 |
0.0897904714 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 10 |
Hb_000661_120 |
0.0907448238 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 11 |
Hb_000003_400 |
0.0909520952 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_004324_330 |
0.0913850142 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 13 |
Hb_033642_120 |
0.0932751994 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 14 |
Hb_000795_040 |
0.0939455608 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 15 |
Hb_000175_410 |
0.0955223149 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_011972_020 |
0.0965252511 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_057688_010 |
0.0968808984 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000710_040 |
0.0984390705 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 19 |
Hb_000111_170 |
0.0998544373 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000205_260 |
0.0999427062 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |