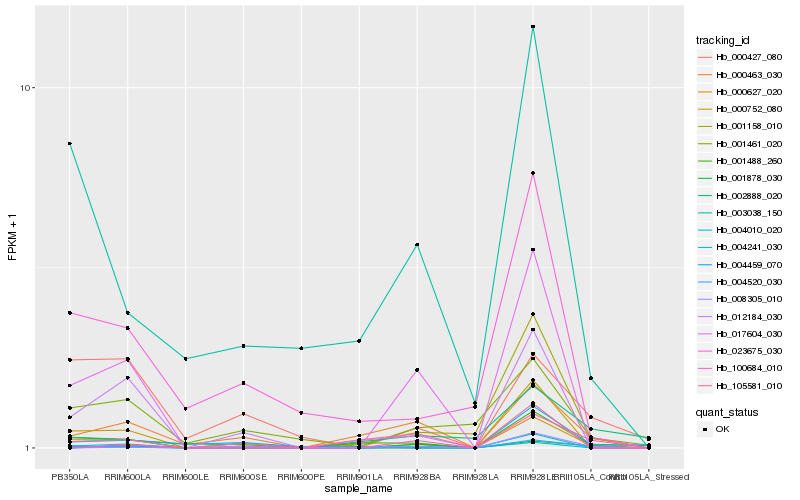
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004520_030 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_000463_030 |
0.2755819842 |
- |
- |
- |
| 3 |
Hb_100684_010 |
0.2883693998 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 4 |
Hb_001488_260 |
0.298387077 |
- |
- |
Alpha-2-macroglobulin [Gossypium arboreum] |
| 5 |
Hb_017604_030 |
0.3024547848 |
- |
- |
- |
| 6 |
Hb_008305_010 |
0.3034916226 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_000752_080 |
0.3052602615 |
- |
- |
- |
| 8 |
Hb_012184_030 |
0.3083964274 |
- |
- |
- |
| 9 |
Hb_004241_030 |
0.3087565456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004459_070 |
0.3262442765 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 11 |
Hb_001158_010 |
0.3271433243 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 12 |
Hb_000627_020 |
0.328943879 |
- |
- |
hypothetical protein MIMGU_mgv1a021461mg, partial [Erythranthe guttata] |
| 13 |
Hb_003038_150 |
0.3299533774 |
- |
- |
- |
| 14 |
Hb_001461_020 |
0.3315274425 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_023675_030 |
0.334193019 |
- |
- |
- |
| 16 |
Hb_001878_030 |
0.3384715051 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105126050 [Populus euphratica] |
| 17 |
Hb_105581_010 |
0.3395380515 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 18 |
Hb_002888_020 |
0.3414284689 |
- |
- |
- |
| 19 |
Hb_004010_020 |
0.3429367352 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 20 |
Hb_000427_080 |
0.3540183608 |
- |
- |
PREDICTED: uncharacterized protein LOC105775598 [Gossypium raimondii] |