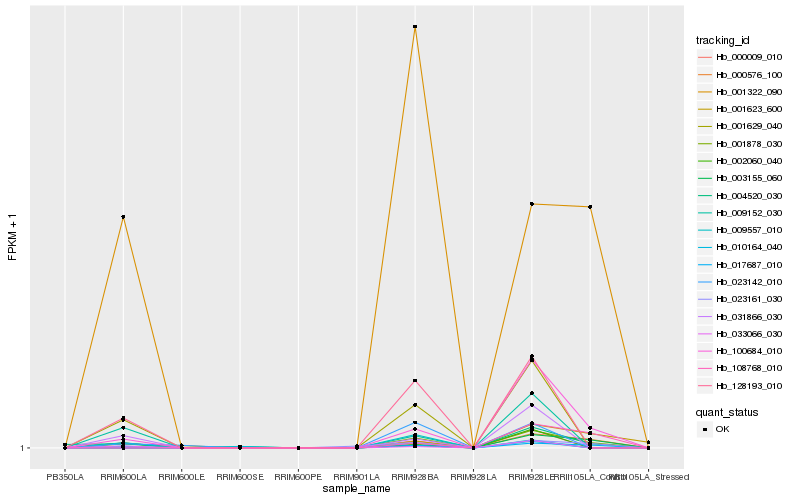
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001878_030 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105126050 [Populus euphratica] |
| 2 |
Hb_100684_010 |
0.1908332954 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 3 |
Hb_001322_090 |
0.2543420254 |
- |
- |
PREDICTED: uncharacterized protein LOC104607608 [Nelumbo nucifera] |
| 4 |
Hb_001629_040 |
0.2753431135 |
- |
- |
hypothetical protein POPTR_0001s41630g [Populus trichocarpa] |
| 5 |
Hb_128193_010 |
0.2760521525 |
- |
- |
PREDICTED: uncharacterized protein LOC103699534, partial [Phoenix dactylifera] |
| 6 |
Hb_017687_010 |
0.2795277784 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy subclass [Oryza sativa Japonica Group] |
| 7 |
Hb_000009_010 |
0.2809228653 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 8 |
Hb_031866_030 |
0.3200989031 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis melo] |
| 9 |
Hb_009152_030 |
0.3252874689 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 10 |
Hb_002060_040 |
0.32566336 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 11 |
Hb_033066_030 |
0.325976237 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 12 |
Hb_001623_600 |
0.3300326379 |
- |
- |
PREDICTED: uncharacterized protein LOC104748784 [Camelina sativa] |
| 13 |
Hb_004520_030 |
0.3384715051 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_023142_010 |
0.3407009861 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_009557_010 |
0.3425157624 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_108768_010 |
0.3456708597 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 17 |
Hb_000576_100 |
0.345849412 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 18 |
Hb_003155_060 |
0.3458648209 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 19 |
Hb_023161_030 |
0.3460998617 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 20 |
Hb_010164_040 |
0.3492774271 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |