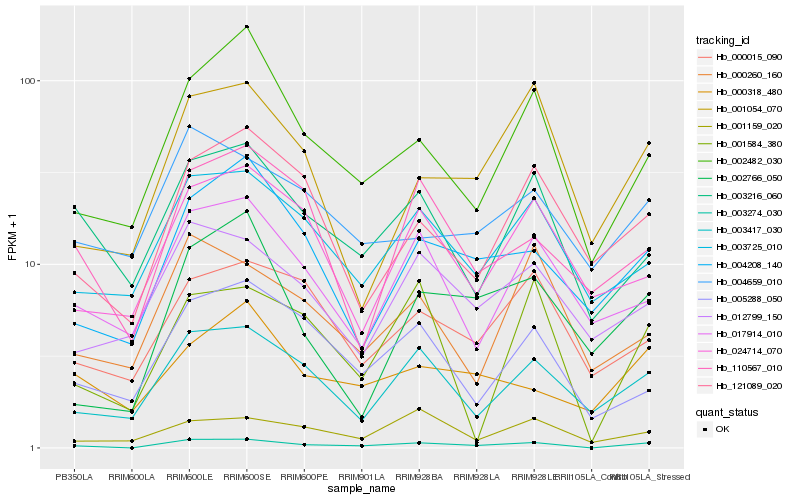
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003274_030 |
0.0 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 2 |
Hb_003417_030 |
0.2036863362 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002482_030 |
0.2120283798 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 4 |
Hb_003725_010 |
0.2142377211 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 5 |
Hb_003216_060 |
0.2173129949 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 6 |
Hb_004208_140 |
0.2216794644 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 7 |
Hb_012799_150 |
0.2253889473 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methyl-CpG-binding domain-containing protein 13 [Jatropha curcas] |
| 8 |
Hb_001584_380 |
0.2275121881 |
- |
- |
PREDICTED: tropomyosin-like [Jatropha curcas] |
| 9 |
Hb_005288_050 |
0.2275414245 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 10 |
Hb_000015_090 |
0.2277822531 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 11 |
Hb_121089_020 |
0.2288850061 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 12 |
Hb_001054_070 |
0.2289690703 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 13 |
Hb_017914_010 |
0.2298881236 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 14 |
Hb_000318_480 |
0.2303395817 |
- |
- |
PREDICTED: RING-H2 finger protein ATL65 [Jatropha curcas] |
| 15 |
Hb_001159_020 |
0.2338255705 |
- |
- |
- |
| 16 |
Hb_004659_010 |
0.2351657059 |
- |
- |
Guanine nucleotide-binding subunit beta-2 [Gossypium arboreum] |
| 17 |
Hb_002766_050 |
0.2355242079 |
- |
- |
PREDICTED: probable carboxylesterase 16 [Jatropha curcas] |
| 18 |
Hb_110567_010 |
0.2363165887 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_024714_070 |
0.2364486046 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 20 |
Hb_000260_160 |
0.2370751612 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |