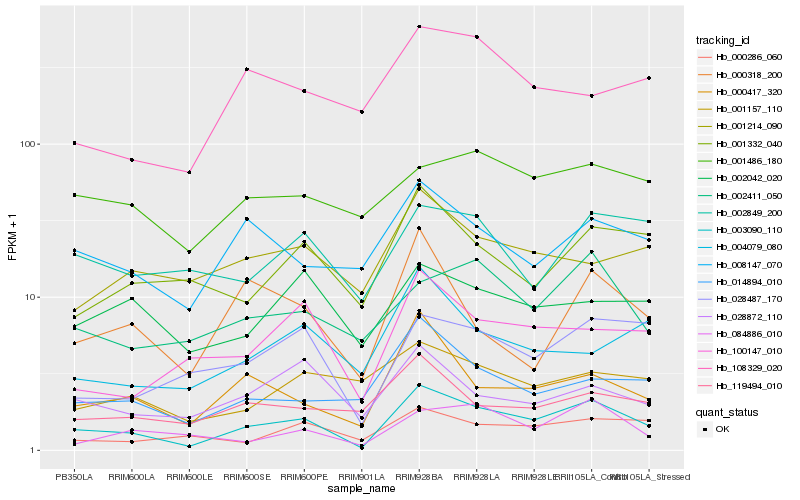
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003090_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000417_320 |
0.1851308519 |
- |
- |
hypothetical protein CICLE_v10000333mg [Citrus clementina] |
| 3 |
Hb_028487_170 |
0.1946615591 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 4 |
Hb_014894_010 |
0.1956806384 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 5 |
Hb_100147_010 |
0.1978159196 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 6 |
Hb_008147_070 |
0.2006773089 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 7 |
Hb_001332_040 |
0.2038935384 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 8 |
Hb_028872_110 |
0.2081218587 |
- |
- |
PREDICTED: uncharacterized protein LOC105644776 [Jatropha curcas] |
| 9 |
Hb_119494_010 |
0.2082718834 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 10 |
Hb_000286_060 |
0.208464366 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 11 |
Hb_002411_050 |
0.2084767165 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000318_200 |
0.2086531757 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 13 |
Hb_084886_010 |
0.21393293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002849_200 |
0.2152597383 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 15 |
Hb_004079_080 |
0.2167103648 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 16 |
Hb_001214_090 |
0.2189469055 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 17 |
Hb_002042_020 |
0.2192226088 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 18 |
Hb_108329_020 |
0.2197962827 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 19 |
Hb_001157_110 |
0.221561463 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 20 |
Hb_001486_180 |
0.2220809081 |
- |
- |
hypothetical protein B456_007G260200 [Gossypium raimondii] |