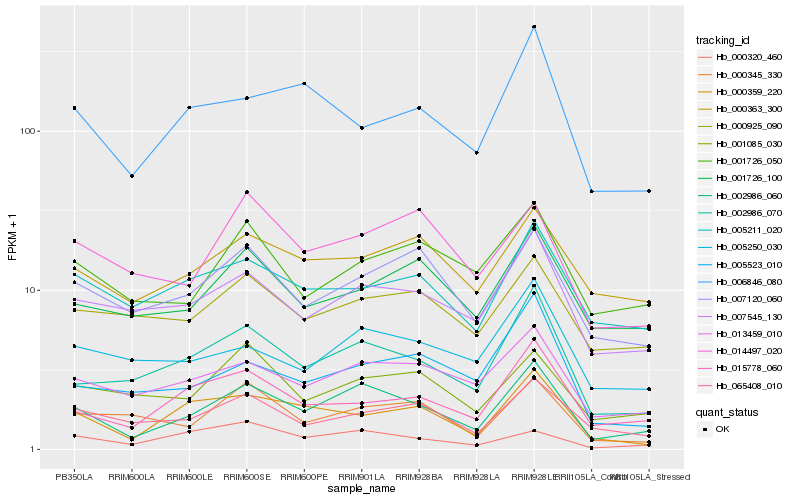
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002986_060 |
0.0 |
- |
- |
hypothetical protein M569_15879, partial [Genlisea aurea] |
| 2 |
Hb_002986_070 |
0.1268196354 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 3 |
Hb_013459_010 |
0.1439691242 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 4 |
Hb_007545_130 |
0.1510212298 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_015778_060 |
0.1555932093 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 6 |
Hb_007120_060 |
0.1601712487 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 7 |
Hb_000359_220 |
0.1617798829 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 8 |
Hb_065408_010 |
0.1623170395 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001085_030 |
0.1692398385 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 10 |
Hb_001726_100 |
0.1692420194 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000345_330 |
0.16971463 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance-like protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_005250_030 |
0.1699101792 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 13 |
Hb_001726_050 |
0.1707651536 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 14 |
Hb_000363_300 |
0.1720527303 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005211_020 |
0.1723062545 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_005523_010 |
0.1724684798 |
transcription factor |
TF Family: PHD |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 17 |
Hb_006846_080 |
0.1735135839 |
- |
- |
calnexin, putative [Ricinus communis] |
| 18 |
Hb_000320_460 |
0.1746407789 |
- |
- |
PREDICTED: uncharacterized protein LOC105650098 [Jatropha curcas] |
| 19 |
Hb_014497_020 |
0.1750873502 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 20 |
Hb_000925_090 |
0.1753145544 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |