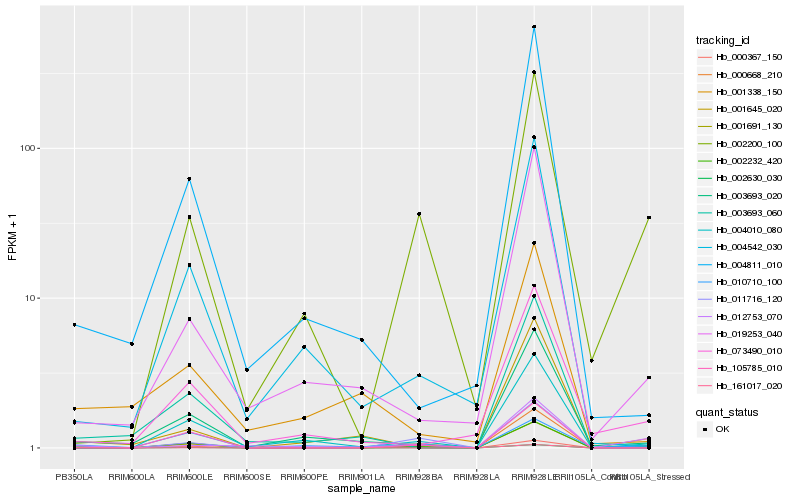
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002232_420 |
0.0 |
- |
- |
Palmitoyltransferase TIP1, putative [Ricinus communis] |
| 2 |
Hb_003693_020 |
0.1999011114 |
- |
- |
Ycf1, partial (chloroplast) [Clusia rosea] |
| 3 |
Hb_004010_080 |
0.2208070733 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_073490_010 |
0.2279447129 |
- |
- |
NAD(P)-linked oxidoreductase superfamily protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_019253_040 |
0.2318741833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_012753_070 |
0.2344920844 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 7 |
Hb_002630_030 |
0.2360017185 |
- |
- |
OSIGBa0132I10.1 [Oryza sativa Indica Group] |
| 8 |
Hb_161017_020 |
0.238914597 |
- |
- |
hypothetical protein EUTSA_v10012231mg, partial [Eutrema salsugineum] |
| 9 |
Hb_011716_120 |
0.2395338247 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 10 |
Hb_001691_130 |
0.2408275605 |
- |
- |
PREDICTED: uncharacterized protein LOC104587431 [Nelumbo nucifera] |
| 11 |
Hb_001338_150 |
0.2409272689 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 12 |
Hb_001645_020 |
0.2409568802 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit H [Medicago truncatula] |
| 13 |
Hb_002200_100 |
0.2421459066 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 14 |
Hb_004811_010 |
0.2435297844 |
- |
- |
DNA-directed RNA polymerase subunit beta, partial [Medicago truncatula] |
| 15 |
Hb_010710_100 |
0.2459659581 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_004542_030 |
0.2460093289 |
- |
- |
hypothetical protein AMTR_s00334p00014280 [Amborella trichopoda] |
| 17 |
Hb_003693_060 |
0.2464046456 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |
| 18 |
Hb_000367_150 |
0.2468351193 |
- |
- |
- |
| 19 |
Hb_105785_010 |
0.2485695697 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 20 |
Hb_000668_210 |
0.2490819354 |
- |
- |
hypothetical protein PHAVU_006G031500g [Phaseolus vulgaris] |