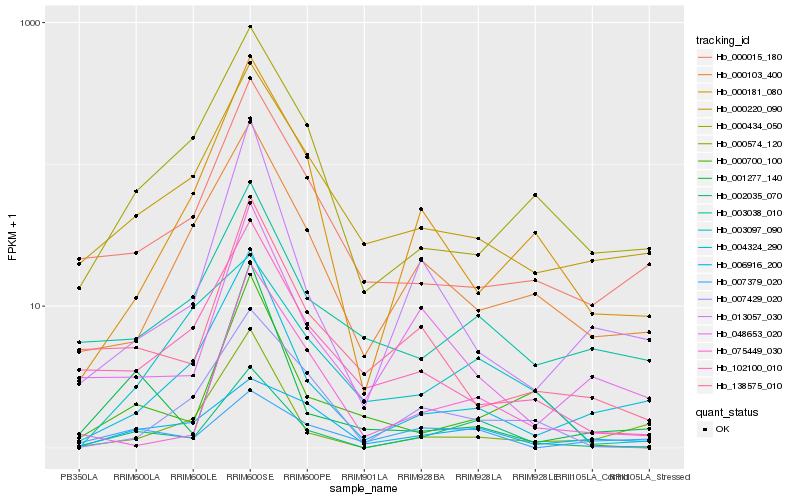
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002035_070 |
0.0 |
- |
- |
hypothetical protein CICLE_v10033569mg [Citrus clementina] |
| 2 |
Hb_003038_010 |
0.2443922666 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000220_090 |
0.2505409285 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 4 |
Hb_007429_020 |
0.2555618024 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_004324_290 |
0.257648574 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 6 |
Hb_048653_020 |
0.2579157548 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000700_100 |
0.2602007348 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 8 |
Hb_075449_030 |
0.2619798914 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 9 |
Hb_000103_400 |
0.2622225717 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000434_050 |
0.2638816643 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 11 |
Hb_000181_080 |
0.264219036 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 12 |
Hb_102100_010 |
0.2660801182 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 13 |
Hb_000015_180 |
0.2668824192 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 14 |
Hb_006916_200 |
0.2688595351 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 15 |
Hb_138575_010 |
0.2702429149 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_013057_030 |
0.2726028036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003097_090 |
0.2761431424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007379_020 |
0.2773061253 |
- |
- |
hypothetical protein JCGZ_25958 [Jatropha curcas] |
| 19 |
Hb_000574_120 |
0.2787290101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001277_140 |
0.279569808 |
- |
- |
PREDICTED: plasma membrane calcium-transporting ATPase 2-like [Takifugu rubripes] |