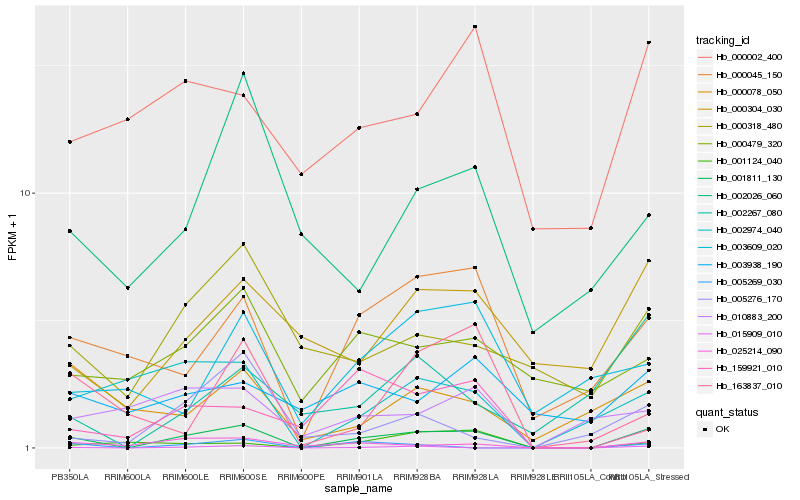
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001811_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_025214_090 |
0.288980766 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 3 |
Hb_000045_150 |
0.2925839482 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 4 |
Hb_015909_010 |
0.3108049441 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_005269_030 |
0.3298189844 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 6 |
Hb_002974_040 |
0.3325909842 |
- |
- |
GRF zinc finger containing protein [Medicago truncatula] |
| 7 |
Hb_000304_030 |
0.3357522556 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 8 |
Hb_000318_480 |
0.3410771217 |
- |
- |
PREDICTED: RING-H2 finger protein ATL65 [Jatropha curcas] |
| 9 |
Hb_163837_010 |
0.3426031276 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Eucalyptus grandis] |
| 10 |
Hb_000002_400 |
0.3445447295 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor 2 [Jatropha curcas] |
| 11 |
Hb_002026_060 |
0.3450737391 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 12 |
Hb_000078_050 |
0.3475075028 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82 [Jatropha curcas] |
| 13 |
Hb_000479_320 |
0.3493300943 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06140, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_159921_010 |
0.3505678095 |
- |
- |
- |
| 15 |
Hb_005276_170 |
0.3512911481 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010883_200 |
0.3516677518 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 17 |
Hb_001124_040 |
0.3527401836 |
- |
- |
PREDICTED: uncharacterized protein LOC103413595 [Malus domestica] |
| 18 |
Hb_002267_080 |
0.353057002 |
- |
- |
PREDICTED: uncharacterized protein LOC105635094 [Jatropha curcas] |
| 19 |
Hb_003938_190 |
0.3549036747 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06145-like [Jatropha curcas] |
| 20 |
Hb_003609_020 |
0.3557064207 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |