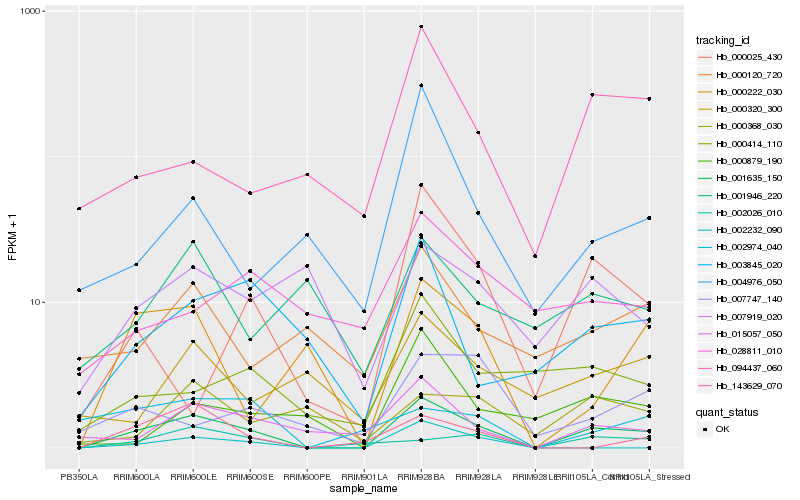
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001635_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_24809 [Jatropha curcas] |
| 2 |
Hb_015057_050 |
0.2939364718 |
- |
- |
D-alanine aminotransferase, putative [Ricinus communis] |
| 3 |
Hb_002026_010 |
0.3075465617 |
transcription factor |
TF Family: mTERF |
hypothetical protein CICLE_v10020375mg [Citrus clementina] |
| 4 |
Hb_003845_020 |
0.3109965697 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_143629_070 |
0.3192232521 |
- |
- |
PREDICTED: uncharacterized protein LOC105647357 [Jatropha curcas] |
| 6 |
Hb_000222_030 |
0.3259741103 |
- |
- |
- |
| 7 |
Hb_000414_110 |
0.3303074533 |
- |
- |
PREDICTED: MATE efflux family protein 5 isoform X1 [Populus euphratica] |
| 8 |
Hb_094437_060 |
0.3320419979 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |
| 9 |
Hb_002232_090 |
0.3328569925 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 10 |
Hb_000025_430 |
0.3346814274 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000879_190 |
0.3373690821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000120_720 |
0.3409042124 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 13 |
Hb_007919_020 |
0.345774765 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 14 |
Hb_000320_300 |
0.3470579974 |
- |
- |
prephenate dehydratase, putative [Ricinus communis] |
| 15 |
Hb_004976_050 |
0.3471257568 |
- |
- |
- |
| 16 |
Hb_007747_140 |
0.3473724479 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_002974_040 |
0.3517902638 |
- |
- |
GRF zinc finger containing protein [Medicago truncatula] |
| 18 |
Hb_001946_220 |
0.3520855163 |
- |
- |
PREDICTED: signal peptidase complex catalytic subunit SEC11A-like [Jatropha curcas] |
| 19 |
Hb_000368_030 |
0.353358752 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 20 |
Hb_028811_010 |
0.3546727044 |
- |
- |
PREDICTED: uncharacterized protein LOC105650178 isoform X1 [Jatropha curcas] |