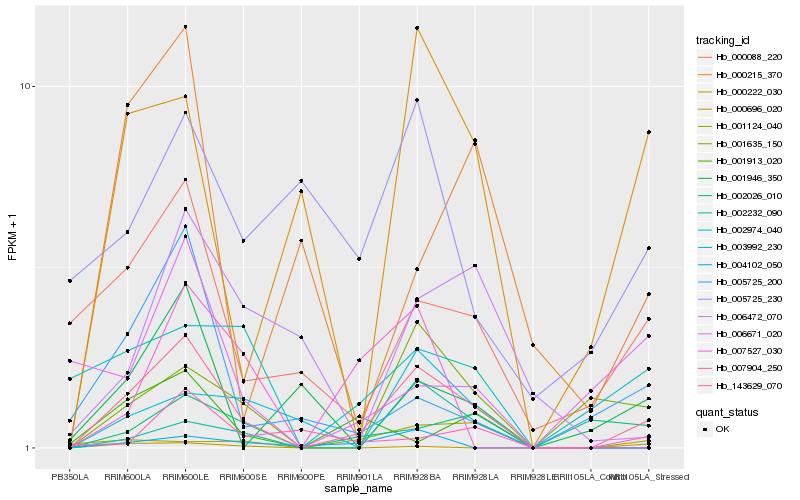
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143629_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647357 [Jatropha curcas] |
| 2 |
Hb_001635_150 |
0.3192232521 |
- |
- |
hypothetical protein JCGZ_24809 [Jatropha curcas] |
| 3 |
Hb_000088_220 |
0.3243907273 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_002026_010 |
0.3439453623 |
transcription factor |
TF Family: mTERF |
hypothetical protein CICLE_v10020375mg [Citrus clementina] |
| 5 |
Hb_000222_030 |
0.3451619605 |
- |
- |
- |
| 6 |
Hb_002974_040 |
0.3459131763 |
- |
- |
GRF zinc finger containing protein [Medicago truncatula] |
| 7 |
Hb_005725_200 |
0.3524505099 |
transcription factor |
TF Family: GRF |
hypothetical protein JCGZ_15989 [Jatropha curcas] |
| 8 |
Hb_002232_090 |
0.3661150671 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 9 |
Hb_000215_370 |
0.37309143 |
- |
- |
myoinositol oxygenase, putative [Ricinus communis] |
| 10 |
Hb_006671_020 |
0.3730966932 |
- |
- |
hypothetical protein CISIN_1g0407181mg, partial [Citrus sinensis] |
| 11 |
Hb_003992_230 |
0.3776531713 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 12 |
Hb_001946_350 |
0.3777385117 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 13 |
Hb_006472_070 |
0.3786528586 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 14 |
Hb_005725_230 |
0.3787353206 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 15 |
Hb_000696_020 |
0.3804269826 |
- |
- |
hypothetical protein JCGZ_25344 [Jatropha curcas] |
| 16 |
Hb_004102_050 |
0.3810997363 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 17 |
Hb_007904_250 |
0.3824073223 |
- |
- |
PREDICTED: vignain-like [Jatropha curcas] |
| 18 |
Hb_007527_030 |
0.3831844718 |
- |
- |
- |
| 19 |
Hb_001913_020 |
0.3871852492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001124_040 |
0.3895925214 |
- |
- |
PREDICTED: uncharacterized protein LOC103413595 [Malus domestica] |